Advances in the use of molecular tools in ecological and biodiversity assessment of aquatic ecosystems
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
Limnetica, 39(1): 419-440 (2020). DOI: 10.23818/limn.39.27 © Asociación Ibérica de Limnología, Madrid. Spain. ISSN: 0213-8409 Advances in the use of molecular tools in ecological and biodiversity assessment of aquatic ecosystems Maria João Feio1,*, Ana Filipa Filipe2,3, Aina Garcia-Raventós2,3, Alba Ardura4, Ana Raquel Calapez1,5, Ana Maria Pujante6, Andreia Mortágua7, Cesc Múrria8, Daniel Diaz-de-Quijano9, Filipa MS Martins2,10, Sofia Duarte11, Marta Sáinz Bariáin12, Rita Cordeiro13, Sinziana F. Rivera14, Leif O. S. Väisänen15, Amélia Fonseca13, Vítor Gonçalves13, Eva Garcia-Vazquez4, David Vieites Rodríguez16, Elena A. Ivanova9, Filipe O. Costa11, José Barquín12, Verónica Rojo17, Joaquín Vierna17, Maria Fais11, Marcos Suarez4, Marko Nieminen18, Monica Ham- mers-Wirtz19, Olesia V. Kolmakova20, Maria Y. Trusova20, Pedro Beja2,3, Raquel González6, Serge Planes21 and Salomé FP Almeida7 1 Marine and Environmental Sciences Centre - MARE & University of Coimbra, Faculty of Sciences and Technology, Department of Life Sciences, Coimbra, Portugal. 2 CIBIO/InBio, Centro de Investigação em Biodiversidade e Recursos Genéticos, Universidade do Porto, Campus de Vairão, Vila do Conde, Portugal. 3 CIBIO/InBio, Centro de Investigação em Biodiversidade e Recursos Genéticos, Instituto Superior de Agronomia, Universidade de Lisboa, Lisboa, Portugal. 4 Department of Functional Biology, University of Oviedo, C/ Julian Claveria s/n 33006-Oviedo, Spain. 5 Linking Landscape, Environment, Agriculture and Food – LEAF, School of Agriculture, University of Lisbon, Lisbon, Portugal. 6 Laboratorios Tecnológicos de Levante, S.L., Avda. Benjamin Franklin, 16, 46980 Paterna, Valencia, Spain. 7 Department of Biology and GeoBioTec – GeoBioSciences, GeoTechnologies and GeoEngineering Research Centre, University of Aveiro, Campus de Santiago, 3810-193, Aveiro, Portugal. 8 Grup de Recerca Freshwater Ecology, Hydrology and Management (FEHM) and Institut de Recerca de la Biodiversitat (IRBio), Departament de Biologia Evolutiva, Ecologia i Ciències Ambientals, Facultat de Biologia, Universitat de Barcelona, Avinguda Diagonal 643, 08028 Barcelona, Catalonia, Spain. 9 Siberian Federal University, Faculty of Biology and Biotechnology, Department of Aquatic and Terrestrial Ecosystems, Svobodnyy 79, 660041 Krasnoyarsk, Russian Federation. 10 Departamento de Biologia, Faculdade de Ciências da Universidade do Porto, Porto, Portugal. 11 Centre of Molecular and Environmental Biology (CBMA), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. 12 Environmental Hydraulics Institute, University of Cantabria, C/ Isabel Torres 15, Parque Científico y Tecnológico de Cantabria, 39011 Santander, Spain. 13 CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, InBIO Laboratório Associado, Pólo dos Açores, Portugal, Faculdade de Ciências e Tecnologia, Universidade dos Açores, 9501-801 Ponta Delgada, Portugal. 14 CARRTEL, INRA, Université de Savoie Mont Blanc, 75 Av. de Corzent, 74200 Thonon les bains Cedex, France. 15 Stream Ecology Research Group, Department of Ecology and Genetics, University of Oulu, Finland. 16 National Museum of Natural Sciences, Spanish National Research Council (CSIC), Calle José Gutiérrez Abascal 2, 28006 Madrid, Spain. 17 Allgenetics, Edificio CICA, Campus de Elviña, s/n, E-15008 A Coruña, Spain. 18 FAUNATICA, Kutojantie 11, Espoo, Finland. 19 Research Institute for Ecosystem Analysis and Assessment, Kackertstr. 10, 52072 Aachen, Germany. 20 Biophysics Institute of the Siberian Branch of the Russian Academy of Sciences (BI SB RAN), 50, Akadem Gorodok, Str. 50, 660036 Krasnoyarsk, Russia.
420 Feio et al. 21 EPHE-UPVD-CNRS, Université de Perpignan, 52 Avenue Paul Alduy, 66860 Perpignan Cedex, France & Laboratoire d’Excellence “Corail”, CRIOBE, BP 1013, 98 729 Papetoai, Moorea, French Polynesia. * Corresponding author: mjf@ci.uc.pt Received: 28/11/18 Accepted: 09/09/19 ABSTRACT Advances in the use of molecular tools in ecological and biodiversity assessment of aquatic ecosystems Conservation and sustainable management of aquatic ecosystems is a priority in environmental programs worldwide. However, these aims are highly dependent on the efficiency, accuracy and cost of existent methods for the detection of keystone species and monitoring of biological communities. Rapid advances in eDNA, barcoding and metabarcoding promoted by high-throughput sequencing technologies are generating millions of sequences in a fast way, with a promising cost reduction, and overcoming some difficulties of the traditional taxonomic approaches. This paper provides an updated broad perspective of the current developments in this dynamic field presented in the special session (SS) “The use of molecular tools in ecological and biodiversity assessment of aquatic ecosystems” of the XIX Congress of the Iberian Association of Limnology (AIL2018), held in Coimbra, Portugal. Developments presented are mainly focused on the Iberian Peninsula (Portugal and Spain, including Atlantic Macaronesian islands) but include studies in France, Germany, Finland, Russia (Siberia) and South America. The networks within which these researchers are involved are yet even broader, profiting from existing molecular facilities, and traditional taxonomic expertise, which can be viewed as a characteristic of this new research area. It was evident in the SS that the use of molecular tools is widespread, being used to study a diversity of aquatic systems, from rivers’ headwaters to estuaries and coastal lagoons, and volcanic, mountain and frozen lakes to hot springs. The organisms targeted are likewise varied and include fish, macroin- vertebrates, meiofauna, microalgae such as diatoms and dinoflagellates, other protists, fungi, and bacteria (cyanobacteria and other). Some studies address the whole biodiversity (i.e., all species present independently of the taxonomic group) from environmental samples of water, biofilms and preservative solution from field samples (e.g., ethanol from macroinvertebrate samples). Great advances were acknowledged in the special session, namely in the use of metabarcoding for detecting hidden biodiversity, juvenile stages, low-abundance species, non-indigenous species and toxicity potential, and ultimately for ecologi- cal monitoring of diatoms and invertebrates. Yet, several drawbacks were highlighted and need further work, which include: taxonomic gaps in the reference databases (including gaps at species level and on intraspecific variability) or absence of public databases (e.g. for meiofauna), still high sequencing costs, the need of a substantial bioinformatics effort, difficulties in establishing the amount of environmental sample necessary for a good DNA extraction and the need for testing different genetic markers to obtain accurate results. Key words: eDNA, metabarcoding, conservation, ecological quality, species detection, rivers, lakes, thermal springs, estuaries, lagoons RESUMO Avanços no uso de ferramentas moleculares na avaliação ecológica e biodiversidade dos ecossistemas aquáticos A conservação e gestão sustentável dos ecossistemas aquáticos é uma prioridade nos programas ambientais em todo o mundo. No entanto, esses objetivos são altamente dependentes da eficiência, precisão e custo dos métodos existentes para detectar espécies e monitorizar comunidades biológicas. Avanços recentes no que respeita ao ADN ambiental e ‘barcoding’ e ‘meta- barcoding’, promovidos por tecnologias de sequenciação designadas ‘high-throughput sequencing’, têm gerado milhões de sequências de forma rápida, com uma promissora redução de custos num futuro próximo, e superando algumas dificuldades das abordagens taxonómicas tradicionais. Este artigo vem fornecer uma perspetiva atualizada e abrangente dos desenvolvi- mentos neste campo que foram apresentados na sessão especial (SE) “O uso de ferramentas moleculares na avaliação ecológi- ca e da biodiversidade dos ecossistemas aquáticos”, no XIX Congresso da Associação Ibérica de Limnologia (AIL2018) realizado em Coimbra, Portugal. Os desenvolvimentos apresentados centram-se principalmente na Península Ibérica (Portugal e Espanha, incluindo as ilhas atlânticas), mas também em França, Alemanha, Finlândia e Rússia (Sibéria). No entanto, as redes em que estes investigadores estão envolvidos são ainda mais amplas, aproveitando as infraestruturas moleculares e o conhecimento taxonómico existentes. Ficou claro na SE que o uso de ferramentas moleculares está disseminado, sendo usado numa diversidade de sistemas aquáti- cos, desde as cabeceiras dos rios aos estuários e lagoas costeiras, e desde lagos vulcânicos, de montanha e congelados, a fontes Limnetica, 39(1): 419-440 (2020)
Molecular tools for aquatic ecosystems’ assessment 421
termais. Os organismos estudados são também variados e incluem peixes, macroinvertebrados, meiofauna, microalgas tal como
diatomáceas e dinoflagelados, outros protistas, fungos e bactérias (cianobactérias e outros). Alguns estudos abordam toda a
biodiversidade a partir de amostras ambientais de água, biofilmes e solução conservante. Grandes avanços foram reconhecidos
na sessão especial, nomeadamente no uso de ‘metabarcoding’ para a deteção de biodiversidade críptica, estádios juvenis,
espécies de reduzida abundância, espécies não nativas, do potencial de toxicidade e, finalmente, para a monitorização ecológica
de diatomáceas e invertebrados. No entanto, dificuldades também foram assinaladas, que necessitarão de mais investimento
futuro, e que incluem: lacunas taxonómicas das bibliotecas de referência (incluindo ao nível da espécie e da intra-variabilidade
de espécies), ausência de bibliotecas públicas (por exemplo, para meiofauna), altos custos de sequenciação, a necessidade de
um esforço substancial de bioinformática, dificuldades em estabelecer a quantidade de amostra ambiental necessária para uma
boa extração de DNA e a necessidade de testar diferentes marcadores genéticos para obter resultados precisos.
Palavras chave: eDNA, metabarcoding, conservação, qualidade ecológica, detecção de espécies, rios, lagos, fontes termais,
estuários, lagoas
INTRODUCTION of disturbances. Therefore, it is essential to
discover and understand the biodiversity present
Biological diversity means the variability among in a certain study area, which is a challenging task.
living organisms from all sources including Most of the traditional approaches for assessing
terrestrial, marine and other aquatic ecosystems biodiversity, where species are identified based on
and ecological complexes of which they are part; their morphological characters, are time-consum-
this includes diversity within species, between ing, expensive and require high taxonomic exper-
species and of ecosystems (Wilcox, 1984). Biodi- tise (Leese et al., 2016). On the other hand, rapid
versity reflects the ecosystem’s health and re- assessment based on an estimation of the abun-
silience to withstand and recover from a variety dance and distribution of target species through
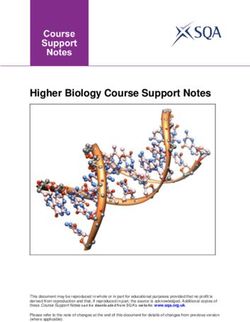
Figure 1. Distribution of the contributors to the special session “The use of molecular tools in ecological and biodiversity assessment
of aquatic ecosystems” of the XIX Congress of the Iberian Association of Limnology (AIL2018) in the World. Image produced in
Google Maps (2019). Distribuição dos autores da sessão especial “O uso das ferramentas moleculares na avaliação ecológica e
biodiversidade dos ecossistemas aquáticos”, do XIX Congresso da Associação Ibérica de Limnologia (AIL2018) no mundo. Imagem
produzida no Google Maps (2019).
Limnetica, 39(1): 419-440 (2020)422 Feio et al.
molecular tools may be conducted in a short time a more accurate species detection and distribution
more cheaply and easily (Minchin et al., 2016). in a specific ecosystem. This is the case of the
For instance, using species-specific DNA mark- meiofauna, which comprises organisms between
ers, the presence of one target species from water 30-1000 µm (Higgins & Thiel, 1988). Due to
samples can be detected using PCR and simple their small size, morphotaxonomic inventories
electrophoresis in agarose gel. This is an efficient can largely fail to identify accurately (Alves et
and convenient approach when the target species al., 2015). Various taxonomic meiofaunal groups
is known because it is a reproducible, fast and a of an estuary in the North of Portugal have been
cost-efficient method (Ardura et al., 2015a; Clusa detected by a target region (Fais et al., AIL2018).
et al., 2016; Devloo-Delva et al., 2016; Ardura et Phytoplankton and general microeukaryotic
al., AIL2018). plankton dynamics under the formation of
The special session “The use of molecular ice-and-snow cover were studied in a Siberian
tools in ecological and biodiversity assessment of mountain lake through molecular techniques
aquatic ecosystems” of AIL2018 (XIX Iberian (Díaz-Quijano et al., AIL2018).
Association of Limnology meeting in Coimbra, Other examples of detection of small organ-
Portugal, June 2018) aimed to present and discuss isms are the microalgae dinoflagellates or
recent studies undertaken in the Iberian Peninsula, diatoms, which have additionally high morpho-
other European countries and South America, in logical similarities and lack of unique characteris-
order to promote knowledge exchange and envis- tics between different species (Lin et al., 2009).
age on future research directions in this area. The The eDNA (environmental DNA) analysis has been
authors represented 13 countries and 46 institu- used in French coastal lagoons to detect a set of
tions (including research institutions, official signal species using mitochondrial cytochrome
agencies and companies), which highlights the oxidase I gene (COI), such as, 21 genera of Dino-
fast development of this area around the world and flagellates and 9 genera of diatoms, including
the importance of broad networks in the advance- Chaetoceros and Nitzschia involved in harmful
ment of this particular research field (Fig. 1). algal blooms (HABs); and invasive invertebrate
The different ways of using molecular species (barnacles, copepods, polychaeta and
approaches in the context of ecological and biodi- ascidians), some of them being pollution indicators
versity assessment in aquatic ecosystems highlight- (Polydora cornuta, Ficopomatus enigmaticus and
ed in the studies presented in the SS (Table 1) were Hydroides elegans) (Ardura et al., AIL2018).
synthesized in the section “Perspectives on the use Ecological impact of algal toxicity is also
of molecular tools.” From those studies, we extract- being investigated through molecular tools
ed the main contributions for the area (section (Cordeiro et al., AIL2018). Toxins are transferred
“Main findings”), as well as the main problems and along the food chain, from different microalgae
gaps identified by the researchers (section “Main (mainly Dinoflagellates, Cyanobacteria, and
drawbacks”) and ended with general inferences and Diatoms) and HABs can be responsible for
future research directions (section “Conclusions”). massive fish mortality (Thangaraja et al., 2007),
while the presence of toxins in fish or shellfish
PERSPECTIVES ON THE USE OF MOLE- can cause severe human diseases (e.g., diarrheic
CULAR TOOLS shellfish poisoning). In the Azorean archipelago
(Portugal), the potential for cyanotoxin produc-
I. Improvement of biodiversity detection and tion was assessed in thermal environments and
biological quality monitoring with molecular freshwater lakes, which are common in these
tools volcanic islands. The confirmation of cyanobac-
teria’s DNA and potential risk of cyanotoxin
Biodiversity production in the eDNA samples (Cordeiro et al.,
AIL2018), revealed to be an efficient method for
Molecular tools are particularly useful to assess monitoring these ecosystems and help to prevent
the diversity of concealed communities, allowing threats to public and environmental health (Pear-
Limnetica, 39(1): 419-440 (2020)Molecular tools for aquatic ecosystems’ assessment 423
son & Neilan, 2008; Salmaso et al., 2017). al., 2019). This approach requires following the
Genetic tools have been increasingly used for Water Framework Directive (WFD; European
studying invasions, because it allows species Union 2000) sampling protocols but avoids the
identification (e.g. Ardura et al., 2010; Ardura & sorting step of separating animals in a sample from
Planes, 2017), determination of the region of vegetation, sediment and litter, which is very
origin (Ardura et al., 2013) and time of initial time-consuming. The authors are examining the
incursion of non-indigenous species (Hilbish et performance of different laboratory procedures on
al., 2000; Rius et al., 2014; Teske et al., 2014). species detection based on the preservative liquid,
This is especially important as the number of and compared taxa recovery with the conventional
introduced species has been increasing during the morphological method. More than half of the taxa
last decades, in freshwater ecosystems (Elvira & found in ethanol were macroinvertebrates targeted
Almodóvar, 2001; Anastácio et al., 2018). One by WFD, while the remaining percentage was
example is the minnow species (Phoxinus genus), identified as, e.g., bacteria, Stramenopiles, terres-
a freshwater fish that has been used as live bait trial invertebrates, amphibians and fishes (Martins
since the 1900s. Individuals were sampled in the et al., AIL2018; Martins et al., 2019).
Douro basin (Portugal) and morphologically
identified as Phoxinus bigerri, a common Biological quality monitoring
minnow in the Iberian Peninsula. Nevertheless,
barcoding showed that the population caught The use of molecular tools in biological quality
closer to the Atlantic Ocean is phylogenetically monitoring is becoming more and more realistic
closer to Phoxinus phoxinus from Charente river and several studies highlighted its potential (e.g.,
in France, confirming for the first time the Filipe et al., 2018; Filipe et al., AIL2018). Com-
presence of this species in the Douro basin (Gar- parison between morphology and metabarcod-
cia-Raventós et al., AIL2018). ing-based approaches to determine species com-
Apart from the tools used for single and position at estuarine sites indicated that species
mixed-organism samples, other sources of DNA richness, one of the metrics frequently used in
have been explored for faster biodiversity assess- bioassessment, would be considerably underesti-
ment such as, DNA from sediment samples, mated if only morphological methods were used
water or sample preservation liquids (e.g., Ayla- (Lobo et al., 2017a).
gas et al., 2016; Deiner et al., 2017; Hajibabaei et In the ecological quality assessment of rivers,
al., 2012). These approaches avoid the traditional diatoms are one of the obligatory elements,
sampling protocols that require a large invest- according to the WFD. Thus, a considerable
ment in human resources with many specialists effort has been made to develop diatom metabar-
studying different biological elements. In these coding and optimize different stages of the
cases, DNA is extracted directly from environ- process (choice of primers, Kermarrec et al.,
mental samples (e.g., water) followed by 2014; diatom barcode database, Rimet et al.,
high-throughput sequencing (HTS) metabarcod- 2016; DNA extraction, Vasselon et al., 2017a;
ing. Taking into account previous results of DNA quantification bias, Vasselon et al., 2018; bioin-
extraction directly from the water (Ardura et al., formatics treatment, Coissac et al., 2012). In
2015a; Zaiko et al., 2015; Ardura & Planes, France, diatom metabarcoding has been applied
2017) a HTS tool was developed to obtain a base- successfully at small (80 samples, Vasselon et al.,
line of biodiversity from 10 different coastal 2017b; Rivera et al., 2020) and larger monitoring
lagoons (Ardura et al., AIL2018). networks (447 samples). In rivers of Central
Alternatively, Martins and collaborators (CIBI- Portugal, the comparison between the Portuguese
O/InBIO University of Porto, Aqualogus company official monitoring index for diatoms (IPS –
and Polytechnic Institute of Bragança) are explor- Indice de Polluosensibilité Spécifique), calculated
ing the option of DNA metabarcoding from based on morphological identification data and on
preservative ethanol of freshwater macroinverte- Operational Taxonomic Units (OTUs) converted
brate samples (Martins et al., AIL2018; Martins et into species data, showed a high correlation
Limnetica, 39(1): 419-440 (2020)424 Feio et al.
Table 1. Biological groups, water bodies and barcode genes assessed in studies presented in the special session “The use of molecu-
lar tools in ecological and biodiversity assessment of aquatic ecosystems” of the XIX Congress of the Iberian Association of
Limnology (AIL2018). Grupos biológicos, massas de água e barcodes analisados nos estudos apresentados na sessão especial ”O
uso das ferramentas moleculares na avaliação ecológica e biodiversidade dos ecossistemas aquáticos”, do XIX Congresso da
Associação Ibérica de Limnologia (AIL2018).
Biological group Type of water Barcode gene Reference
body/location
Total biodiversity – eDNA Coastal lagoons of COI, 18S Ardura et al.,
(water) Gulf of Lyon - AIL2018
France
Total biodiversity – eDNA Rivers and estuaries COI, 18S, Sainz-Bariáin et
(water, sediment) – Pas, Asón, Miera 16S al., AIL2018
rivers (Cantabria),
Douro, Ebro
Fish Rivers – Douro 12S – MiFish Filipe et al.,
catchment region AIL2018
Fish (non-indigenenous species) Rivers - Douro COI, Cytb Garcia-Raventós
– Phoxinus phoxinus catchment et al., AIL2018
Macroinvertebrates and eDNA Rivers – Tua (Douro COI Martins et al.,
(ethanol) catchment) AIL2018
Macroinvertebrates Rivers – Spain 16S Pujante et al.,
(Mediterranean AIL2018
rivers), Finland and
Germany
Macroinvertebrates and eDNA Rivers – Lobregat, COI Múrria et al.,
(water) (Mediterranean river, AIL2018
Catalonia)
Diatoms Rivers – central rbcL Mortágua et al.,
Portugal AIL2018;
Mortágua et al.,
2019
Diatoms Lakes – Bourget, rcbL Rivera et al.,
France 2018; Rivera et
al. AIL2018
Biofilms (bacteria, fungi, Rivers (mesocosms) 16S, 18S Calapez et al.,
microalgae) AIL2018; Calapez
et al., 2019
Phytoplankton Mountain lake - 18S Díaz-Quijano et
Oiskoe, Siberia al., AIL2018
Algae (toxicity, Cyanobacteria) Thermal waters and 16S and sxtA, Cordeiro et al.,
freshwater lakes – sxtI, sxtH, AIL2018
Azores islands sxtG for
saxitoxinas,
anaC, anaF
for anatoxina,
and mcyC,
mcyD, mcyE,
mcyG for
microcistina
Meiofauna (sediment) Estuary – Lima river, COI, 18S Fais et al.,
Portugal AIL2018
Limnetica, 39(1): 419-440 (2020)Molecular tools for aquatic ecosystems’ assessment 425
(Mortágua et al., AIL2018). Besides, more than drift sampling (intervals of 1 hour). Results
half (ca. 56 %) of the samples shared the same showed that while the traditional and bulk
water quality class either using the conventional sampling approaches detected essentially riverine
or the molecular approach. These results show the species, the eDNA also captured terrestrial asso-
potential for adaptation of present taxonomic ciated fauna (Múrria et al., AIL2018).
indices to molecular data, as it was concluded in Development of indices based on molecular
studies in Mayotte island, France (Vasselon et al., information for the monitoring of aquatic ecosys-
2017b) and in the UK (Kelly et al., 2018). tems (i.e., ecological status or conservation
The benthic invertebrates are another compul- status) is the purpose of the work developed at the
sory quality element of the WFD. In Portugal, five University of Cantabria. Yet here, the main goal
sites sampled in Tua river (Douro basin) were is a global assessment of water bodies through
classified to the same quality status through both eDNA from water and sediment (Sainz-Bariáin et
morphological identification and ethanol-based al., AIL2018). Additionally, the study of bacterial
DNA metabarcoding (Martins et al., AIL2018; diversity and primary producers through
Martins et al., 2019) when applying the Iberian metagenomics is aimed, which could give com-
Biological Monitoring Working Party (IBMWP) plementary information on ecosystem functions
index with presence/absence data, at family level (e.g., organic matter degradation or primary
(Alba-Tercedor et al., 2002). However, only about production under different conditions).
half of the species identified by metabarcoding Molecular analysis constitutes, in addition, a
were detected by morphology, whereas the former simpler way of analysing the impact of anthropo-
missed about 20 % of the species identified genic and natural alterations in complex commu-
morphologically, corresponding to taxa with a low nities composed of microorganisms. A study in
frequency (< 5 individuals). mesocosms run by Calapez and collaborators
In Valencia, the Laboratorios Tecnológicos de (Universities of Aveiro and Coimbra, Portugal)
Levante (Pujante et al., AIL2018) in the context analysed stream biofilm responses to multi-
of the European project BIOWAT-KIT ple-stressors typical of Mediterranean streams and
(DNA-based kit for biodiversity assessments and found biofilm community shifts induced by flow
biomonitoring of European water bodies), are stagnation, organic loads and grazing activity.
developing and validating a genomic tool for the Specifically, the OTUs determination helped to
identification and assessment of diversity of investigate how biofilm microbial communities’
benthic invertebrate communities in Europe, with proportions changed under the different stressor
the aim of improving and facilitating the combinations more quickly. The interaction of
bioassessment. An audit (made by taxonomists) those three stressors altered algae, fungi and
to an official European freshwater monitoring bacteria diversity proportions within the biofilm,
program, based on macroinvertebrate samples, with a synergistic effect on fungal diversity, while
revealed that 29-30 % of the specimens had been algae and bacteria had an antagonistic response to
overlooked by the primary taxonomists (Haase et stressors’ interaction (Calapez et al., AIL2018).
al., 2010). For 16 % of the samples, these discrep-
ancies led to different final ecological assessment II. Molecular analysis in aquatic water bodies
and demonstrated the need for adequate quality
control and auditing in freshwater monitoring. Different aquatic systems have been studied
Múrria and collaborators (University of Barcelo- through molecular techniques by the teams
na, Spain and Salford, Manchester, UK) used present in the SS: rivers and streams, lakes,
metabarcoding techniques to compare the thermal waters and estuaries and coastal lagoons.
estimates of the ecological status using traditional
morpho-taxonomy against high-throughput DNA Rivers and streams
sequencing of: 1) bulk sampling (after sorting
individuals from multi-habitat Surber samples), Rivers of NW Iberian Peninsula (Portugal and
2) eDNA (water samples) and 3) invertebrate Spain) have been studied under the FRESHING
Limnetica, 39(1): 419-440 (2020)426 Feio et al.
project (Next-generation biomonitoring: freshwa- and are currently being identified (Sainz-Bariáin
ter bioassessment and species conservation et al., AIL2018).
improved with metagenomics) by CIBIO/InBIO,
covering up to 150 sampling sites (Filipe et al., Lakes
AIL2018). Each site was sampled using conven-
tional methods along with water sampling from The studies presented in the SS addressed a wide
different microhabitats in order to maximize the diversity of freshwater lakes. The Azores archipel-
detection of several taxa present in the water body ago (Portugal) located in the North Atlantic Ocean
through eDNA. However, results shown in the is composed of nine islands, which are very impor-
special session focused on freshwater fish. In tant and unique in terms of biodiversity, climate,
central Portugal, the studies of the University of volcanic activity and geomorphology (Antunes &
Coimbra and Aveiro and partners from INRA Rodrigues, 2011). Fifteen freshwater lakes from
Thonon, France, include 88 sites located in the the Archipelago of the Azores in São Miguel, Pico,
catchments of rivers Vouga, Mondego and Lis in Flores and Corvo islands were studied to investi-
a total area of 11 215 km2. These sites were gate cyanotoxin production potential.
sampled for algae and macroinvertebrates, but In France, diatom metabarcoding has been
present results report to diatoms only (Mortágua applied to assess the structure of diatom commu-
et al., AIL2018; Mortágua et al., 2019). nity and the ecological status of the littoral zone
In the BIOWAT-KIT project, three rivers of Lake Bourget (deepest French lake). The struc-
from each country (Spain, Finland and Germany) ture of the assemblages based on the morphologi-
have been selected to test a genomic tool across cal (taxa lists) and molecular (OTUs lists) identi-
different European regions covering a variety of fication of diatoms were well correlated. Howev-
climatic and geomorphological conditions. In er, the ecological status of the lake varied
Spain, the rivers are typically Mediterranean with between these two methods since floristic inven-
different characteristics: Júcar is a calcareous tories differed significantly (Rivera et al., 2018;
mountain river; Mijares is a low-mountain river Rivera et al., AIL2018). The main reason for this
with high mineralization; while the Turia river is discrepancy was the incompleteness of the
a low altitude river (Pujante et al., AIL2018). diatom reference database (Diat.barcode, former-
Another Mediterranean river from Catalonia, the ly called R-Syst::diatom in Rimet et al., 2016).
Llobregat (156 km in length), was studied by In Cantabria, five mountain lakes were
Múrria and collaborators, which covers a gradient sampled for molecular analysis of environmental
of pollution and anthropogenic impact. This is a samples (water and sediment). The first is located
well-studied river (Munné & Prat, 2004, 2011), at ca. 1870 m of altitude in the Liordes Valley, a
which includes a pollution gradient from pristine unique ecosystem in the Picos de Europa massif,
headwater reach, through site located down- located in a glacial-karst depression surrounded
stream of a big reservoir or salt mining, to urban by calcareous walls. The Lloroza lakes (ca. 1800
and agricultural landscapes at lowlands. A m of altitude) are small lagoons of karstic nature
sampling of macroinvertebrates was done in 5 located in Picos de Europa National Park in the
sites along the river (Múrria et al., AIL2018). Cantabria province. Finally, the Enol and Ercina
In Cantabria, two rivers, Pas and Asón (at ca. 110m of altitude) are two glacial lakes
(Spain), with temperate hyper-oceanic climate forming the Covadonga lakes located within the
with sub-Mediterranean characteristics were Picos de Europa National Park in the Asturias
studied with molecular tools to compare diversity province. These samples are still being processed
under pristine and polluted conditions. In addi- (Sainz-Bariáin et al., AIL2018).
tion, water and biofilm samples were recently In Siberia, the Oiskoe mountain lake is being
collected from 96 river sites belonging to the studied with phytoplankton samples through
Douro, Ebro and Cantabrian basins (Spain). metabarcoding from a conservation perspective
These sites were sampled to determine the total (Diaz-de-Quijano et al., AIL2018). Located in the
biodiversity from microorganisms to vertebrates Ergaki Natural Park, West Sayan Mountains, is a
Limnetica, 39(1): 419-440 (2020)Molecular tools for aquatic ecosystems’ assessment 427
poorly studied area due to its extreme climate with Minho (Portugal) team is monitoring meiofauna
a wide range of annual temperatures (-41 °C to communities of this estuary through metabarcod-
+32 °C). The lake is surrounded by a mosaic land- ing, annually, whose preliminary results were
scape of bogs, sparse taiga forest, scree and alpine presented at the AIL conference (Fais et al., 2018,
tundra and biodiversity has particular adaptations AIL2018).
to these conditions (Anishchenko et al., 2015). In Cantabria, five sediment and five water
However, human activities, namely tourism and samples were taken from 3 estuaries (Pas, Miera,
global warming in South Siberia and Central Asia, and Asón) characterized by large intertidal
are the present threats to these ecosystems. surfaces and dominated by the tidal dynamic,
making them well-mixed estuaries. This coast is
Thermal waters subjected to various anthropogenic pressures.
These sites have been sampled to determine
In São Miguel island, in the Azorean archipelago, general biodiversity through molecular analysis.
environmental samples were collected from 21 The team from the University of Oviedo
thermal sites, including hot springs, thermal pools (Spain) has been using metabarcoding (eDNA) to
and ponds, thermal streams and hydrothermal determine the biodiversity and detect particular
vents, with temperatures ranging from 28 °C to organisms in the coastal lagoons of Gulf of Lyon,
over 90 °C (Cordeiro et al., AIL2018). Cyanobac- in the French Mediterranean coast (Ardura et al.,
teria were isolated from these samples and depos- AIL2018). Ten lagoons were analysed: Berre,
ited in BACA-Banco de Algas e Cianobactérias Beaduc, Bages-Sigean, La Palme, Leucate,
dos Açores (Universidade dos Açores), which is Mejean, Prevost, Thau, Vic and Canet. These
part of REBECA (Red de excelencia en biotec- ecosystems provide habitat for many species,
nología azul (algas) de la región de la Macarone- nursery areas and feeding grounds for marine and
sia). From the 40 strains isolated, 24 strains and estuarine fish (Perez-Ruzafa et al., 2011). They
environmental samples were targeted for cyano- support important fisheries and allow for inten-
toxin production potential through conventional sive aquaculture exploitation (Cataudella et al.,
PCR. Preliminary results show that none of the 2015). Despite most of them being under protec-
studied cyanobacteria strains have cyanotoxin tion, they still suffer from several threats derived
production potential (Cordeiro et al., AIL2018). from human activities such as pollution, eutrophi-
cation, climate change and introduction of
Estuaries and coastal lagoons non-native species (≈100 non-indigenous species
were identified; Reizipoulou et al., 1996; Chap-
Finally, studies have been undertaken in estuaries man, 2012).
and coastal lagoons. A proof-of-concept study
(Lobo et al., 2017b) on the application of DNA III. Selection of adequate barcode genes for
metabarcoding for monitoring estuarine macro- each group of organisms
zoobenthic communities has been conducted in
the Sado estuary (SW Portugal). The metabarcod- The selection of barcode genes varies with the
ing approach was able to discriminate macrozoo- target taxonomic group studied and the focus of
benthic communities among sampling sites the studies. The researchers took different options
successfully and provided biotic index levels in the studies presented in the SS:
comparable to the morphology-based approach
(Lobo et al., 2017b). Up north, in river Lima (NW COI
Portugal), the estuarine area has become an
important Portuguese harbour, used for commer- The DNA barcode region elected most frequently
cial navigation and fishing activities and is for the identification of individualized specimens
subjected to constant dredging as well as the input is a fragment of the mitochondrial COI gene
of agricultural run-off and urban and industrial (Herbert et al., 2003). The Cytochrome c (COI) is
sewage (Sousa et al., 2007). The University of an amino acid sequence that is highly conserved
Limnetica, 39(1): 419-440 (2020)428 Feio et al.
in eukaryotes, differing by only a few residues. ers (e.g., Borges et al., 2012). In the Lima estuary,
There are robust universal primers for it that DNA from meiofauna communities was extract-
recover most animal phyla, and thousands of ed from intertidal sediments. In this case, the
reference sequences are available in public target genes were the COI and 18S ribosomal
databases such as BOLD and GenBank (Ratnas- RNA (18S rDNA) gene. MiSeq amplicon
ingham & Hebert, 2013; Hebert et al., 2013). sequences were processed in mothur (version
However, the high variability in the third position 1.39.5, Schloss et al., 2009) by using appropriate
of the COI codons makes it difficult to design bioinformatic procedures; while the taxonomy of
universal primers for metabarcoding DNA the processed sequences were assessed by blast-
studies (Ficetola et al., 2010). For fish identifica- ing against the full ntNCBI database (Fais et al.,
tion, most used barcode markers in DNA refer- AIL2018). This database was chosen due to the
ence collections are the COI and cytochrome b lack of adequate reference sequences in
(Cytb) genes, other mitochondrial genes, which better-known databases, such as BOLD (Ratnas-
can confirm taxonomic identification at the ingham & Hebert, 2007) and Silva (Pruesse et al.,
species level. However, some studies are showing 2007). In the French coastal lagoons, the inverte-
that COI might not be the best option for assess- brate communities were as well analysed from
ing and monitoring freshwater fish diversity eDNA with COI marker.
using environmental DNA from water because
this marker might not contain suitably conserved 18S, rbcL and 16S
regions (e.g., Deagle et al., 2014). Instead, the
potential of using the MiFish region from the In the project BIOWAT-KIT a preliminary evalu-
ribosomal 12S is under consideration (Miya et ation of different genomic regions using publicly
al., 2015; Filipe et al., AIL2018). available sequence data was carried out in order
For Iberian freshwater macroinvertebrates, to identify the best-suited DNA barcode marker
public repositories for COI DNA barcodes cover for the identification of 141 families of inverte-
35 % of the taxa (3348 morphospecies) (Múrria et brates belonging to four different phyla (Platyhel-
al., AIL2018, Múrria et al., 2020). However, this minthes, Annelida, Mollusca, and Arthropoda).
coverage is highly variable across taxonomic Several primer pairs have been designed, includ-
groups. For instance, Odonata (79 species, 54.43 ing a degenerate primer pair and a cocktail of
%), Hemiptera (81 species, 54.32 %), Mollusca group-specific primers, which will presumably
(65 species, 53.85 %), Trichoptera (390 species, amplify all the target invertebrate taxa present in
50.77 %) and Crustacea (10 species, 50.5 %) freshwater samples. Based on the results, the
were the best-represented groups, whereas mitochondrial 16S gene was selected for the
Diptera (1693 species, 23.21 %), and Plecoptera DNA metabarcoding analysis of freshwater
(135 species, 31.11 %) were the less barcoded invertebrate communities within this project,
orders. Portuguese invertebrate communities since it combines both conserved regions suitable
sampled were also processed for metabarcoding for primer design, and variable regions with good
using a small COI fragment (313bp) by Martins taxonomic resolution at the family level (and
and collaborators in CIBIO (AIL2018; Martins et potentially, also at the genus or species level)
al., 2019). The HTS data were identified against (Pujante et al., AIL2018).
the invertebrate collection of the InBIO Barcod- In Thonon (France), the INRA team targeted
ing Initiative (at CIBIO-UP) that includes several genes for diatoms (18S, COI, rbcL) (Ker-
hundreds of specimens of macroinvertebrate taxa marrec et al., 2013). While COI is found in mito-
from northeast Portugal. chondrial DNA of eukaryotic organisms, the 18S
Macroinvertebrates and marine fish have been is part of the ribosomal RNA of eukaryotes and
the target of comprehensive DNA barcoding the ribulose-1,5-bisphosphate carboxylase/oxy-
campaigns across multiple coastal ecosystems in genase (rcbL) is present in plants chloroplasts.
continental Portugal. The primary marker was the The rcbL showed to be the most suitable barcode
COI, occasionally supplemented by other mark- for biomonitoring purposes with diatoms (Ker-
Limnetica, 39(1): 419-440 (2020)Molecular tools for aquatic ecosystems’ assessment 429
marrec et al., 2013; Kermarrec et al., 2014; Paw- (DGGE) was run for each community, conducted
lowski et al., 2016). Thus, DNA metabarcoding in a DCode system (Bio-Rad, Hercules, CA,
of periphytic diatom community samples from USA). DGGE images were converted, normal-
Portuguese and French rivers included a step for ized, and analysed with the software BioNumer-
DNA extraction using commercial kit NucleoS- ics 7.6 (Applied Maths, Sint-Martens-Latem,
pin® Soil and a second step for DNA sequencing Belgium) to obtain the relative abundances
with MiSeq system (Illumina) using rbcL plastid according to gel band intensity (OTUs).
gene (312 bp barcode) (Mortágua et al., In the Russian lake Oiskoe, planktonic
AIL2018, Mortágua et al., 2019, Rivera et al., microeukaryotes were assessed before and after
2018; Rivera et al., 2020). Sample sequences ice-and-snow cover formation (Díaz-Quijano et
obtained from metabarcoding were then analysed al., AIL2018). The focus was set on phytoplank-
using the software mothur (version 1.39.5, ton and general protists, but other eukaryotic
Schloss et al., 2009). Taxonomic assignment of actors of the microbial loop, such as ciliates and
OTUs was based on the Diat.barcode library fungi were assessed as well. General eukaryote
(available at: https://www6.inra.fr/carrtel- primer pair targeting the V4 region of the small
collection_eng/Barcoding-database). In French subunit 18S rRNA gene was used (Balzano et al.,
lagoons, the process was similar, but the DNA 2015). This is a modification of Stoeck’s primer
extraction was done with the kit Power Water pairs (Stoeck et al., 2010), with an extra degener-
DNA Isolation MOBIO® and sample sequences ate nucleotide position, which allows haptophytes
obtained from metabarcoding were then analysed to be targeted.
using the software QIIME (https://qiime2.org). Total biodiversity (from microorganisms to
In Azorean lakes and thermal springs, DNA vertebrates) has also been addressed in projects
was extracted up to 24h after sample collection, developed in Cantabria, with the addition of 16S
according to the gram-negative bacteria protocol and 18S primers for prokaryotes and eukaryotes
of PureLinkTM Genomic DNA Mini Kit (Invitro- (Bact02 and Euka02 primers, respectively),
gen, Carlsbad, CA, USA), followed by amplifica- besides COI for macroinvertebrates (Sainz-Bariáin
tion of genes targeting 16S rDNA and cyanotox- et al., AIL2018).
ins (Microcystin, Saxitoxin and Anatoxin-a)
using conventional PCR and electrophoresis IV. New database entries
protocols (Cordeiro et al., AIL2018). All proto-
cols used were modified from existing ones avail- Continuous incorporation of data from new or
able in the scientific literature (Ouahid et al., updated biological surveys is essential to develop
2005; Ballot et al., 2010; Ledreux et al., 2010; a good species database (Olenin et al., 2016).
Rantala-Ylinen et al., 2011; Casero et al., 2014). Many of the studies presented in the SS originat-
Biofilms from central Portugal and their ed important new barcode data that fed different
response to multiple stressors in mesocosms were databases.
assessed through their OTUs composition in a
study by Calapez et al. (AIL2018). DNA was Fish and invertebrates
extracted from a portion of the biofilm using
PowerSoil® DNA Isolation Kit (Mobio Laborato- For marine life, core COI reference databases for
ries Inc., Carlsbad, CA, USA), followed by a the most prominent groups of Portuguese and
PCR to amplify rDNA genes for each studied Iberian fish and macroinvertebrates were made
biofilm community, using a Taq DNA polymer- publicly available on BOLD systems. Regarding
ase. The bacterial V3 region of 16S was amplified fish, in addition to the Portuguese marine ichthy-
with the primer pair 338F-GC and 518R, the ITS1 ofauna (Costa et al., 2012), reference databases
region of rDNA of fungi using the primer pair have been generated for the Mediterranean
ITS1F-GC and ITS2 and the 18S gene of eukarya (Landi et al., 2014), the North Sea and British
by using the primer pair Euk1A and Euk516r-GC. Isles species (Knebelsberger et al., 2014). A
Then a Denaturing Gradient Gel Electrophoresis published compilation for all European marine
Limnetica, 39(1): 419-440 (2020)430 Feio et al.
fish species is available as well (Oliveira et al., at the Helmholtz Centre for Polar and Marine
2016). For freshwater fish species, the reference Research in Potsdam, Germany (shuang@awi.de).
database for European species is almost complete
concerning standard DNA barcodes (COI) and V. Multidisciplinary international networks
public data can be found in GenBank and BOLD
databases. However, there is only very limited The metagenomics is an area where extended
12S sequence data available that can be used as a networks tend to be formed in order to easily
reference to taxonomically annotate eDNA tackle all the fields involved, encompassing field-
derived OTUs. Among the invertebrates there are work and sample collection to laboratory proce-
published databases and other scattered DNA dures, taxonomic expertise and molecular analy-
barcode contributions available for annelids, ses. This need is clear in the global distribution of
namely Polychaeta (Lobo et al., 2016; Ravara et authors of the SS (Fig. 1).
al., 2017), for molluscs (Gastropoda: Borges et The University of Minho team (Portugal) has
al., 2016; bivalve woodborers: Borges et al., integrated the Consortium for the Barcode of Life
2012), and for crustaceans (e.g. Amphipoda; (CBOL) from early stages and later the Interna-
Lobo et al., 2017b). tional Barcode of Life (iBOL) and in collabora-
tion with the Museu Nacional de História Natural
Meiofauna e Ciência, Instituto Português do Mar e da
Atmosfera, the Portuguese Institute of Malacolo-
Concerning meiofauna, to the best of our knowl- gy, the research institutes IMAR, CIIMAR and
edge, there are no specific databases. Yet, Tang CNC/Biocant, and the Universities of Guelph
and collaborators (2012) gathered a total of 12 (Canada), Bangor (UK) and Vigo (Spain) works
000 sequences (generated and retrieved from to build core reference databases for marine life.
GenBank) across 55 meiofaunal datasets com- The teams from the Universities of Aveiro
prising 3 taxonomic ranks (15 species complexes, and Coimbra (Portugal) have been working with
26 genera, and 14 higher taxa above the genus INRA at Thonon-les-Bains (France) in the labo-
level, including orders, classes, and phyla), using ratorial treatment of periphytic biofilms, from
either 18S or COI markers. extraction, amplification, sequencing of DNA
and bioinformatic analyses. MARE team is also
Diatoms collaborating with CIBIO (Portugal) for the
assessment of freshwater invertebrate communi-
For diatoms, Diat.barcode library (formely ties and biological quality through DNA. For the
called R-Syst::diatom in Rimet et al., 2016) FRESHING project (CIBIO/InBIO, Portugal)
(https://www6.inra.fr/carrtel-collection_eng/ the laboratory procedures and the HTS (MiSeq
Barcoding-database) and was used in the studies v2, 2x250bp PE) were performed in CIBIO-UP
presented at AIL2018 conducted in Portugal by (Portugal) while fieldwork have been done in
Mortágua et al. (AIL 2018; 2019) and in France by collaboration with the company Aqualogus and
Rivera et al. (AIL 2018). This database is open the taxonomical identification at Instituto
access and contains 18S and rbcL barcodes. In Politécnico de Bragança (Portugal). These
addition, R-Syst::diatom provides information teams, like those from Universities of Minho,
concerning morphological diatom features (e.g., Coimbra and Aveiro (Portugal), Cantabria and
biovolumes, chloroplasts, etc.), ecological features Barcelona (Spain), are part of the larger network
(taxa preference to pollution) and life-forms of the European COST action DNAqua-Net,
(mobility, colony-type). The database is uploaded which among other tasks are tackling problems
and curated every six months. The sequences such as an adaptation of currently used biotic
obtained in the Russian study are not attributed to indices for metabarcoding data.
any taxocenose-specific database but should be Samples from the Cantabrian coast (Spain),
made available to the builders of a cryophyllic Gulf of Lion (South France), Polynesian ports
diatom and green algae ribosomic RNA database and Spanish rivers are being processed in molec-
Limnetica, 39(1): 419-440 (2020)Molecular tools for aquatic ecosystems’ assessment 431
ular facilities of the University of Oviedo. DNA faster processing of samples in the lab for DNA
sequencing will be done at the Massive Sequenc- metabarcoding. However, the results are sensitive
ing Service Unit from the IBBTEC (CSIC - to various laboratory procedures, namely DNA
Universidad de Cantabria – Sodercan). The extraction methods and/or the storage and collec-
University of Barcelona team is currently collab- tion timing of preservative ethanol (Martins et al.,
orating with the University of Salford (UK) and AIL2018).
University of Tromsø (Norway) for sequencing
facilities and bioinformatics. In the Azores, all Ecology and conservation
molecular laboratory work is conducted in the
laboratories of the University of Azores (UAc) Molecular analyses in aquatic ecosystems
and CIBIO. The cyanobacteria cultures were brought not only new information but also new
established and maintained in BACA-Banco de questions. DNA barcoding studies on Portuguese
Algas e Cianobactérias dos Açores (UAc), which marine life have been revealing numerous cases
is part of the REBECA network. The team works of comparatively high intra-specific divergences,
on this topic with the Ecotoxicology team from suggesting the existence of considerable hidden
CIIMAR, University of Porto. In Russia, the diversity and putative cryptic species across
molecular facility used was the Laboratory of diverse marine taxa, including fish and major
Experimental Hydroecology, at the Biophysics groups of invertebrates (Costa et al., 2012, Landi
Institute (Siberian branch of the Russian Acade- et al., 2014, Lobo et al., 2016, 2017a, Oliveira et
my of Sciences). Sequencing (Illumina MiSeq) al., 2016). These findings suggest that popula-
was performed in three facilities: Konstantin V. tions of marine organisms may be much more
Krutovsky lab, at the Sukachev Institute of structured than previously thought, calling for a
Forest; the Centre for Collective Use of the Insti- continuous effort on the description of the hidden
tute of Bioorganic Chemistry, Novosibirsk, diversity and further completion of the reference
Russia; and the company Evrogen (Moscow). databases. In order to improve the efficiency of
amplification of COI barcodes from marine
MAIN FINDINGS macrozoobenthos, Lobo et al. (2013) developed a
new pair of degenerate primers with a broad
The SS showed several interesting results at the scope of amplification success across a phyloge-
technical level but also new insights for the ecolo- netically diverse range of marine metazoan taxa.
gy and conservation of aquatic systems. In the very first study based on molecular data
of freshwater diatom communities in Portugal,
Technical aspects the total number of diatom taxa identified was
125 from 88 river samples which corresponded to
It was found that the choice of the markers to target about 41 % of the number of taxa identified by
particular primer pair can considerably influence using the light microscope (Mortágua et al.,
the metabarcoding-based analyses output. For AIL2018; Mortágua et al., 2019). These results,
estuarine meiofaunal. up to 85 % of the species somewhat unexpected, were in accordance with
constituting a mock community were detected by results registered in studies performed in other
using a combination of 3 primer pairs targeting the countries (Vasselon et al., 2017b; Rivera et al.,
COI region, while only 30 to 60 % were recovered 2018 and Keck et al., 2018). A possible explana-
by using any primer set alone (Hollatz et al., 2017; tion might be the high number of unassigned
Fais et al., AIL218). Also, the amount of starting reads, which is a consequence of the incomplete-
material from the sample for eDNA extraction is ness of the reference database.
critical for a comprehensive assessment of meio- The molecular approach was also found
faunal communities in estuarine ecosystems. important in the detection of new introductions of
The use of preservative ethanol from field fishes and tracking introduction histories, which
samples seems to be a promising solution for can be relevant for designing proper management
macroinvertebrate biodiversity assessment, with plans. It is the case of the species P. phoxinus that
Limnetica, 39(1): 419-440 (2020)432 Feio et al.
was recorded for the first time in the Douro Basin. in the Lima estuary and in the Tua river in Portugal
This species can be easily misidentified as other with benthic invertebrates, where a fair number of
species from the same genus when using only OTUs could not be assigned to phylum or other
morphological identifications in the field. lower taxonomic rank due to the primers used for
The eDNA and metabarcoding approaches targeting the COI region (Mortágua et al., 2019). A
were found efficient to obtain accurate baseline similar issue was reported for the diatoms as previ-
information to be used in conservation planning ously referred, in spite of the large database and
and ongoing management of coastal lagoons in diatom cultures existing in Thonon-les-Bains,
the south of France. Despite their different status INRA, with a high number of unassigned reads (67
of conservation within Natural Parks, Reserves or %). The increase in the number of diatom barcodes
Natura 2000 Network, they are already contami- in reference databases will allow for a complete
nated with non-indigenous species, some of them study of diversity, namely in what concerns to rare
already described as invasive species. taxa. In some cases, databases are not sufficient for
New records of cyanobacteria species presence assigning species and they must be assigned at
were detected in the Azores through molecular genus level; in these cases, previous taxonomic
analyses (Cordeiro et al., AIL2018). In addition, work is necessary. In the French coastal lagoons,
some of the sampled lakes cyanotoxins production only ca. 10 % of reads obtained were identified to
potential was confirmed, mainly associated with the species level and those that could not be
eutrophication and anthropogenic effects, which described to the species level had multiple best
shows the potential of molecular tools for monitor- BLAST hits or the best BLAST hit had no
ing cyanotoxin risk in aquatic systems. species-level information available. In addition,
In Russia, a unique dataset of early winter lake local databases covering intra-specific variability
water microbial communities was produced as are important, especially when geographical barri-
winter dynamics are usually out of the scope of ers can lead to high intra-specific variability (e.g.,
limnological studies in Siberia, due to the harsh Douro River Basin).
fieldwork conditions (Diaz-de-Quijano et al.,
AIL2018). The Cryptomycota clade LKM11, Extraction of eDNA
which was previously found in ice-covered lakes
of Antarctica (Rojas-Jimenez et al., 2017), repre- Protocols need further adjustments and should be
sented up to 6-10 % of the reads in intermediate adapted to the environments and types of samples
and deep layers of the water column of the ice-cov- (e.g., biofilms scrapings or preservative liquid of
ered Oiskoe lake. Metabarcoding of microbial but bulk samples instead of water). eDNA extraction
also macroscopic communities enabled an easier was the biggest setback. This was found through
calculation of phylogenetic diversity metrics, and the development of the work with cyanobacteria as
testing hypotheses on the ecological mechanisms they have a wide range of morphological charac-
governing community assemblages. teristics, like mucilage sheaths (Codd et al., 2017),
that makes DNA extraction more complicated.
MAJOR DRAWBACKS Different methods were tested to improve cell
lysis, like sonication, enzymatic lysis and readjust-
Different technical drawbacks were signalized in ments of temperature and incubation time (Kim et
the SS, in spite of the potential advantages of al., 2009). Similar results were found using ethanol
molecular approaches in biodiversity and ecolog- from the preservation of macroinvertebrate
ical assessment of aquatic ecosystems. samples where different DNA extraction methods
retrieved different species diversity across time.
Taxonomic gaps
Amount of environmental sample
In the SS it was often referred to the existence of
taxonomic gaps in the reference databases when The amount of sample needed for good DNA
considering local fauna. One example is the study extraction can be harder to determine since it
Limnetica, 39(1): 419-440 (2020)You can also read