STANDARDIZATION OF INDUCER-ACTIVATED BROAD HOST RANGE EXPRESSION MODULES: DEBUGGING AND REFACTORING AN ALKANE-RESPONSIVE ALKS/PALKB DEVICE ...
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
Synthetic Biology, 2021, 6(1), 1–11
DOI: https://doi.org/10.1093/synbio/ysab030
Advance access publication date: 17 September 2021
Research Article
Standardization of inducer-activated broad host range
expression modules: debugging and refactoring an
alkane-responsive AlkS/PalkB device
Alejandro Arce-Rodríguez1,2 , Ilaria Benedetti1 , José Manuel Borrero-de Acuña2 , Rafael Silva-Rocha1,† , and Víctor de Lorenzo1,*
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
1
Systems Biology Department, Centro Nacional de Biotecnología-CSIC, Campus de Cantoblanco, Madrid, Spain
2
Institute of Microbiology, Technische Universität Braunschweig, Braunschweig, Germany
†
Present address Ribeirão Preto Medical School (FMRP), University of São Paulo Ribeirão Preto 14049-900 Brazil.
*Corresponding author: E-mail: vdlorenzo@cnb.csic.es
Abstract
Although inducible heterologous expression systems have been available since the birth of recombinant DNA technology, the diversity
of genetic devices and their coming together in the corresponding vectors often result in a lack of reproducibility and interoperability. In
an effort to increase the predictability of expression of genes of interest in a variety of possible bacterial hosts, we propose a composition
standard for debugging and reassembling all regulatory parts that participate in the performance of such devices. As a case study, we
addressed the n-octane and dicyclopropyl ketone-inducible PalkB promoter of the alkane biodegradation pOCT plasmid of Pseudomonas
putida. The standardized expression module included an edited alkS transcription factor divergently expressed and separated from
PalkB by a synthetic buffer segment. The DNA sequence of the alkS gene was modified to alleviate the catabolite repression exerted
by several carbon and nitrogen sources through the Crc/Hfq complex of some hosts. The PalkB promoter and the alkS variants were
then formatted as Standard European Vector Architecture cargoes, and their activity in P. putida was quantified with fluorescent and
luminescent reporters. Despite considerable editing of the DNA sequences involved, the thereby refactored module basically kept the
functioning parameters of the original configuration. The same qualities were inspected when the system was passed to Escherichia
coli and Pseudomonas aeruginosa. We argue that the application of the compositional standard thereby implemented in the AlkS/PalkB
module to other promoter/regulator pairs will enable more complex genetic programming in non-model bacteria.
Key words: SEVA; AlkS; Pseudomonas; Crc; Hfq; cytometry; noise
1. Introduction One step in that direction was the creation in 2013 of the so-called
Expression of genes of interest in hosts (e.g. bacteria) different Standard European Vector Architecture (SEVA; 6), which com-
from their native origin and triggered by an external inducer is prises a large number of standardized antibiotic marker genes,
one of the basis of modern biotechnology (1). A large number of broad host range origins or replication and functional cargoes,
genetic devices to this end have been developed over the years aimed at simplifying genetic programming of a wide variety of
following the pioneering use of the Isopropyl β-D-thiogalactoside bacteria of industrial and environmental interest (7). One type
(IPTG)-inducible lac promoter for expression in Escherichia coli (2). of such cargoes includes expression modules. While the bound-
Typical modules (3) encompass one promoter that is activated or aries of the corresponding DNA segment within the plasmid vector
repressed by a cognate transcription factor (TF; either an activa- or transposon vector frame are well defined in the SEVA for-
tor or a repressor). The regulator then binds the target sequence or mat, the organization of the regulatory elements inside the cargo
changes its activity in a fashion dependent on exposure of cells to has not been standardized yet. In this work, we propose a spe-
a physical (e.g. heat and light) or chemical (e.g. inducer) signal. cific arrangement for such inducible modules that attempts to
This basic scheme is the template for a large number of pop- preserve the inducibility of the TF/promoter pairs usable for het-
ular expression vectors based on a suite of regulator/promoter erologous expression while simplifying the native regulation of the
pairs (1, 4, 5). While they have been useful when the issue was to cognate systems.
express one or few genes at a time in one host, the onset of syn- As a case study for such standardization effort, we have chosen
thetic biology in recent years has multiplied the need of multiple, the regulatory node that controls the expression of the alk genes
regulatable promoters endowed with specific parameters and as for biodegradation of octane borne by the OCT plasmid of the soil
independent as possible of the physiological state of the host (5). bacterium Pseudomonas putida GPo1 (8). In its native arrangement,
Submitted: 26 December 2020; Received (in revised form): 8 September 2021; Accepted: 15 September 2021
© The Author(s) 2021. Published by Oxford University Press.
This is an Open Access article distributed under the terms of the Creative Commons Attribution-NonCommercial License
(https://creativecommons.org/licenses/by-nc/4.0/), which permits non-commercial re-use, distribution, and reproduction in any medium, provided the original
work is properly cited. For commercial re-use, please contact journals.permissions@oup.com2 Synthetic Biology, 2021, Vol. 6(1), No. 0
two gene clusters are involved in the process. alkST encodes than induction by DCPK—has been eliminated and replaced by
the transcriptional regulator of the pathway (AlkS, inducible by non-regulatory DNA sequences, following a defined composition
n-octane) and AlkT (a component of alkane hydroxylase). The standard. As shown below, the resulting inducible expression
second alkBFGHJKL cluster determines the rest of the activities, module keeps the functionality of the wild-type arrangement
which are expressed from the upstream AlkS-dependent promoter with very similar performance parameters. On this basis, we
PalkB (9, 10). Once excised from its native context and assembled advocate the general application of the compositional standard
adjacent to each other in a single DNA segment, the alkS/PalkB used to assemble this device for increasing the reproducibility
pair has been used to develop a number of biosensors for alka- and interoperability of a large number of other devices made
nes as well as heterologous expression vectors (11–15). The last with regulatory parts mined from the genomes of environmental
is facilitated by the use of the gratuitous, soluble inducer dicy- bacteria.
clopropyl ketone (DCPK). However, this simple rearrangement of
functional segments with alkS and PalkB does not eliminate the reg-
ulatory complexity embodied in them. alkS is transcribed through
2. Materials and methods
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
two promoters, PalkS1 and PalkS2 , which are negatively and posi- 2.1 Strains, plasmids and growth conditions
tively regulated, respectively, by AlkS. In addition, translation of Bacterial strains and plasmids used in this work are listed in
AlkS is subject to the post-transcriptional control of the Crc/Hfq Table 1. All P. putida specimens were derived from the reference
complex, which introduces an additional regulatory layer (i.e. isolate P. putida KT2440. E. coli strains DH5α and CC118 were
catabolite repression) in the system (16). Finally, the activity of used as hosts for maintenance of plasmids and used as recip-
AlkS seems to be influenced also by the cytochrome terminal ients of the AlkS/PalkB system as indicated in each case. Unless
oxidase Cyo (17). Such a regulatory density allows the extant sys- indicated otherwise, cells were grown at either 30◦ C (P. putida) or
tem to compute many physiological signals other than the mere 37◦ C (E. coli and P. aeruginosa) in rich LB medium (18) amended,
presence of pathway substrates. Yet, the same intricacy is also where necessary, with 100 µg/ml ampicillin (Ap), 50 µg/ml strep-
a nuisance for the predictability of the gene expression module. tomycin (Sm) and 50 µg/ml chloramphenicol (Cm), to retain
In the work presented below, we have constructed a refactored plasmids as necessary. In the case of P. aeruginosa PAO1 cul-
AlkS/PalkB device in which any known regulatory control—other tures, the concentration of Sm was increased to 500 µg/ml. For
Table 1. Strains and plasmids used in this work
Strain/plasmid Description/relevant characteristics Reference
E. coli strains
CC118 F-, ∆(ara-leu)7697, araD139, ∆(lac)X74, phoA∆20, galE, galK, thi, rpsE, rpoB, argE(Am), recA1 (49)
DH5α F-, supE44, ∆lacU169, (ϕ80 lacZDM15), hsdR17, (rk-mk+), recA1, endA1, thi1, gyrA, relA (50)
HB101 SmR , hsdR− M+ , pro, leu, thi, recA (18)
P. putida strains
KT2440 Prototrophic, wild-type strain derived from P. putida mt-2 without pWW0 plasmid (51)
KT2440 crc::Gm P. putida KT2440 with the chromosomal crc gene interrupted with a GmR cassette (16)
P. aeruginosa strain
PAO1 Prototrophic, wild-type reference strain (52)
Plasmids
pRK600 CmR ; oriV ColE1, tra+ mob+ of RK2; helper plasmid for mobilization in tripartite (53)
conjugations
pMA Cloning vector for synthetic DNA GeneArta
pAlkS3 ApR ; pMA cloning vector bearing the optimized sequence of alkS gene This work
pBAM1 Mini-Tn5 suicide delivery vector, source of Pneo promoter (26)
pSEVA429 crc+ SmR , oriRK2, oriT; pSEVA421-derivative carrying an alkSED /PalkB expression system. The gene This work
alkS is edited for SEVA-incompatible restriction sites, but encodes the wild-type primary
amino acid sequence of the AlkS protein and keeps the Crc/Hfq-binding sequence in the
cognate transcript
pSEVA429b SmR , oriRK2, oriT; pSEVA 421-derivative carrying the alkSCR /PalkB expression system. alkS This work
sequence is same as alkSED (Supplementary Figure S1) but 5′ -end edited for removing the
Crc/Hfq binding sequence of the corresponding mRNA
pSEVA421 SmR , oriV RK2, oriT, standard MCS (6)
pSEVA426 SmR , oriV RK2, oriT, luxCDABE reporter system (6)
pSEVA429 → luxCDABE SmR , oriRK2, oriT; pSEVA 429 cloned with the luxCDABE reporter system This work
pSEVA427 SmR , oriV RK2, oriT, green fluorescent protein (GFP) reporter system (6)
pSEVA429 crc+ → GFP pSEVA429 crc+ with a promoterless GFP gene as a transcriptional reporter This work
pSEVA429 crc+ → luxCDABE pSEVA429 crc+ with a promoterless luxCDABE operon as a transcriptional reporter This work
pSEVA429 → GFP pSEVA429 with a promoterless GFP gene as a transcriptional reporter This work
pJAMA30 ApR , oriV ColE1; native PalkST -alkST/PalkB expression system driving the transcription of GFP (12)
tir. This segment is flanked by NotI sites
pARalkS SmR , oriV RK2, oriT; pSEVA421 cloned with the NotI fragment from pJAMA30 carrying the This work
native alkane/DCPK-responsive reporter system
a
https://www.thermofisher.com/content/dam/LifeTech/Documents/geneart/geneart-vector-map.pdf.
b
Complete DNA sequence available in the SEVA database (http://seva-plasmids.com/).A. Arce-Rodríguez et al. 3
solid media preparation, LB medium was supplemented with
1.5% (w/v) Bacto Agar (Pronadisa). Where indicated, the expres-
sion of PalkB promoter was induced by the addition of (DCPK:
Sigma, 95% purity) in solid and liquid media at the concentrations
indicated.
2.2 Recombinant DNA techniques
General methods for DNA manipulation were performed with
standard protocols described elsewhere (18). The amplification of
DNA fragments by polymerase chain reaction (PCR) was imple-
mented in 50-µl reactions containing ∼100 ng of genomic DNA
or 10 ng of plasmid as template, 0.25 mM dNTPs, 25 pmol of
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
each primer and 1 U of GoTaq DNA polymerase (Promega). Reac-
tions were run by an initial denaturalization (5 min, 94◦ C) fol-
lowed by 30 cycles of denaturalization (1 min, 94◦ C), annealing
(1 min, 58◦ C–64◦ C), extension (1–3 min at 72◦ C) and final exten-
sion (10 min, 72◦ C). PCR products were purified with the Nucle-
oSpin® Gel and PCR Clean-up kit (Macherey-Nagel) and, when
required, digested with restriction enzymes purchased from New
England Biolabs. Plasmid DNA was isolated by means of the Wiz-
ard® Plus SV Minipreps DNA Purification system (Promega). E. coli
cells were transformed with plasmids with the CaCl2 method
(18). In the case of P. putida and P. aeruginosa, plasmids were
introduced by either conjugative triparental mating using the
E. coli HB101 (pRK600) as helper strain (19) or electroporation of
cells previously washed with 300 mM sucrose and concentrated in
the same solution (20).
2.3 Construction of the PalkB /AlkS expression
modules compatible with the SEVA
In order to create an expression system that could fit the SEVA
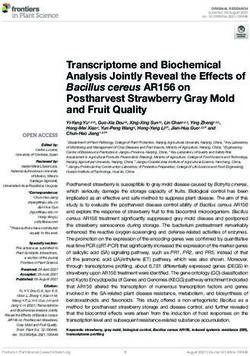
plasmid platform (6, 7), the gene alkS from P. putida GPo1 was Figure 1. Organization of standardized inducible expression modules.
edited to remove incompatible restriction sites (alkSED ). The modi- (A) Arrangement of DNA portion, boundaries and their roles as the
frame for inserting genes for inducer-responsive transcriptional
fied sequence (Supplementary Figure S1) was entirely synthesized
regulators and cognate target promoters. (B) Blowup of the standardized
by GeneArt/Thermo Fisher (Waltham, Massachusetts) and deliv- DNA sequence that acts and the scaffold for the rest of the functional
ered as an insert in the pMA vector that was called pAlkS3. parts. (C) Configuration of reporter plasmids (with GFP or any other gene
Both the AlkS regulatory protein and the PalkB promoter were of interest, GOI) used in this work. Note constant and variable segments.
then arrayed with the other DNA segments indicated in Figure 1.
The PalkB promoter was PCR-amplified with primers aaPalkB1-F
(5′ AGCGGATAACAATTTCACACAGGACGTGTTTTTCCAGCAGACGA template. The forward primer contains the recognition sequence
C3′ ) and aaPalkB1-R (5′ ATGACCTAGGCTCTCGACATCTTAAAC of NcoI enzyme (underlined) and also six single-nucleotide
CTGAGC3′ ), using as template genomic DNA from P. putida PBS4 mutations that eliminate the Crc site from alkS (in bold charac-
(21). The Pneo promoter was also amplified by PCR with oligo- ters). The PCR fragment was then digested with NcoI and XhoI
nucleotides aaPKm-NcoI1-F (5′ TAGAACCATGGTTTTTCCTCCTTA (the last within the alkS sequence), and the resulting 406 bp
TAAAG3′ ; underlined sequence shows Ncol recognition site) and fragment was recloned into the same sites of pSEVA429 crc+ .
aaM13-R24-rev (5′ TCCTGTGTGAAATTGTTATCCGCT 3′ ) from In order to follow the SEVA nomenclature, this final expression
pBAM1 (Table 1). The sequence of primer aaM13-R24-rev is com- vector was named pSEVA429 (i.e. Sm resistant, carrying a RK2 ori-
plementary to the 5′ end of primer aaPalkB1-F (bold characters), gin of replication and AlkS/PalkB expression cargo; Supplementary
allowing the assembly of Pneo with PalkB by SOEing PCR (22). Inser- Figure S2).
tion of the resulting product into the NcoI/AvrII restriction sites
of a pSEVA frame gave rise to expression vector pSEVA429 crc+
2.5 Parameterization of the AlkS/PalkB module
(Supplementary Figure S2).
For generating constructs that report transcriptional activity as a
fluorescent emission, the GFP tir gene of pSEVA427 was cloned into
2.4 Removal of the Crc-binding site in the alkS plasmids pSEVA429 crc+ and pSEVA429 as a HindIII/SpeI insert to
gene generate plasmids pSEVA429 crc+ → GFP and pSEVA429 → GFP,
The Crc-binding site in the 5′ end of the alkS gene in pSEVA429 respectively. As a control, we excised the NotI fragment from
crc+ was modified with PCR-based site-directed mutagenesis. pJAMA30 containing the original alkST genes of pOCT transcribed
Briefly, the first 570 bp of alkS were amplified with the oligonu- by their own PalkST promoter and, in divergent orientation, the
cleotides 5-alkSmut-NcoI (5′ GCGCCCATGGGCATGAAGATCAA PalkB driving the expression of the GFP tir gene (12) (Supplementary
GATCATCAACAATGAT TTCCCGGTTGCCAAGATCG3′ ) and 3-alk Figure S3). This ∼6.7-kb fragment was cloned into the NotI site of
Smut-XhoI (5′ AGCGCCTGCAAGTTTAAGCC3′ ) using the pAlkS3 as pSEVA421 to generate the control vector pARalkS. The three GFP4 Synthetic Biology, 2021, Vol. 6(1), No. 0
tir reporter vectors described above, as well as the promoter-less 96-well assay plate (BD Falcon), regrown to mid-exponential
pSEVA427, were transferred into wild-type P. putida KT2440 cells phase, added with the inducer and luminescence recorded
and into its isogenic crc::Gm derivative. Plasmid-bearing strains after 4 h.
were then grown in LB medium at 30◦ C until mid-exponential
phase, the cultures supplemented with 0.05% v/v DCPK and fluo-
rescent emission for the next 6 h followed with a CyFlow® Cube 2.6 Promoter kinetics and statistical analyses
8 (Partec GmbH, Münster, Germany) flow cytometer. GFP was Promoter induction kinetics were calculated from fluorescence
excited at 488 nm, and the fluorescence signal was recovered with mean values at each timepoint using linear regression analysis.
a 536/40 BP filter. Note that monitoring performance of other These analyses were used to obtain the slope values shown in
expression devices in E. coli (23) and P. putida (24) has been done Table 2. Promoter inducibility was calculated from the data col-
for much longer. Yet, we entertain that the most relevant char- lected 6 h postinduction, using the following equation: (signal
acteristics of the promoter(s) become manifest when cells are ON population−signal promoterless pSEVA427)/(signal OFF pop-
still growing exponentially. This is because transcriptional activ- ulation−signal promoterless pSEVA427). In the sporadic cases
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
ity relies only on the intrinsic characteristics of the engineered where the signal of cells carrying the promoterless pSEVA427
modules before entering stationary phase and become affected was slightly higher than the signal of OFF cells, this parame-
by physiological responses to nutrient starvation. The resulting ter was omitted from the equation. Statistical differences of the
data were processed using FlowJo v. 9.6.2 software (FlowJo LLC, maximum inducibility values between the strains harboring the
Ashland, OR, USA) as follows. Forward and side scatter density different AlkS/PalkB modules were calculated by means of a two-
plots were used to identify the bacterial cell population of inter- tailed Student’s t-test. All statistical analyses were performed with
est and to exclude debris. Data were recorded for at least 10 000 GraphPad Prism v.9.0.1 (GraphPad Software, San Diego, California
bacteria, and the GFP specific output was calculated using popu- USA).
lation mean fluorescence intensities. For the parameterization of
the edited AlkS/PalkB module in other bacterial hosts, E. coli CC118
and P. aeruginosa PAO1 carrying pSEVA429 → GFP were grown in LB
medium at 37◦ C until mid-exponential phase, induced with 0.05%
v/v DCPK and analyzed by flow cytometry as described above. For
monitoring activity of the AlkS/PalkB module at a population level,
plasmids pSEVA429 crc+ and pSEVA429 were inserted with the pro-
moterless luminescent reporter luxCDABE operon excised from
pSEVA426 as a HindIII/SpeI fragment. This originated plasmids
pSEVA429 crc+ → luxCDABE and pSEVA429 → luxCDABE, respec-
tively. As with the GFP counterparts before, these plasmids were
passed to the wild-type P. putida host. For measuring light emis-
sion under various DCPK concentrations, cells grown overnight
in LB were diluted in same medium, placed in a MicrotestTM
Table 2. Promoter expression parameters of the different
AlkS/PalkB modules constructed in this study
Promoter inducibility
Fluorescence (mean fluorescence
Strain/plasmid induction slope (m)a fold change)b
P. putida KT2440
pARAlkS (control) 247.6 ± 6.79 49.37 ± 4.59
pSEVA429 crc+ → 190 ± 4.87 80.53 ± 0.61***
GFP
pSEVA429 → GFP 187.90 ± 4.93 80.98 ± 14.70*
P. putida KT2440 crc::Gm
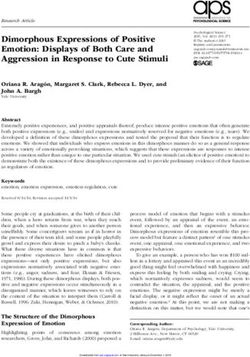
Figure 2. Functional segments of the naturally occurring and
pARAlkS (control) 105.3 ± 4.18 20.22 ± 10.01
standardized AlkS/PalkB device. (A) Native organization of regulatory
pSEVA429 crc+ → 135.2 ± 4.58 59.01 ± 2.86**
parts in the original context of pOCT plasmid. Note complex
GFP arrangement of transcriptional and post-transcriptional signals
pSEVA429 → GFP 139.3 ± 9.738 81.36 ± 6.08*** (e.g. inhibition of alkS mRNA translation by the Crc/Hfq complex) and
E. coli CC118 dual effect of inducer-activated AlkS on self-promoters PalkS1 and PalkS2 .
pSEVA429 → GFP 11.72 ± 0.82 7.07 ± 1.74 (B) Constitution of the standardized AlkS PalkB expression module. The
wild-type DNA sequence of the regulator has been edited to remove
P. aeruginosa PAO1 restriction sites incompatible with the SEVA standard but keeping the
pSEVA429 → GFP 69.98 ± 3.75 27.24 ± 8.15 same primary amino acid sequence (AlkSED ). alkS expression is now
a
under the control of a heterologous Shine-Dalgarno (SD) sequence and a
The slope value was calculated using the linear regression of the induction
constitutive promoter (Pneo , see text) and closer to target, divergent
kinetics as explained in Section 2.6.
b promoter PalkB —albeit separated by the buffer sequence indicated in
Promoter inducibility was calculated 6 h after the first timepoint, as
explained in Section 2.6. The statistical differences between strains harboring Figure 1B. (C) AlkS/PalkB device bearing a regulator variant devoid of its
the pARAlkS control plasmid versus the crc+ /crc− standardized versions were Crc-binding site. As before, this change in the DNA of alkS keeps the
calculated by the Student’s t-test. primary amino acid sequence of the protein identical to the wild-type
*P-values:A. Arce-Rodríguez et al. 5
3. Results and discussion could originate from a poor translation. The segment for consti-
3.1 A compositional standard for engineering tutive expression/translation of the TF is followed upstream by
inducer-dependent heterologous gene expression a 150-bp segment of DNA with no known function or activity
retrieved from the lacIq –Ptrc expression system of plasmid pTrcA
Figure 1A sketches the organization of the inducible expression
module proposed in this work for SEVA cargoes (6) aimed at het- (28). This sequence has no known regulatory elements and it func-
erologous expression of genes of interest in a variety of bacterial tions as a buffer region to ease mutually negative supercoiling
hosts. First, the standard asks for constitutive expression of the that could stem from transcription of divergent promoters (29). As
gene(s) encoding the effector-responsive regulator. In their natu- shown in Figure 1A, the adjacent piece of DNA is the one that bears
ral context, TFs are often subject to a degree of self-regulation, the promoter targeted by the inducible regulator and orientated
either positive or negative (25). This introduces an excessive com- opposite in respect to the sequences for expression of the TF gene.
plexity that the arrangement shown in Figure 1A mitigates if not The specific DNA sequence of this promoter changes from case
entirely eliminates. The standardized sequence that holds the var- to case, but it should by default be accommodated within a seg-
ious functional parts of the device (Figure 1B) is assembled as ment of not more than 100 bp. The 3′ of this promoter sequence
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
follows. The source of transcription of the signal-responsive TF is bound by an AvrII site, which links this segment to the start
gene is the 106 bp minimal promoter P3/Pneo that drives expres- of the SEVA polylinker (6). Note that two sites at the boundary
sion of the kanamycin resistance gene of pBAM1 (26). Follow- buffer sequence/promoter and at the end of the MCS have target
ing this promoter, default translation efficiency is also fixed by sequences for oligonucleotides R24 and F24 (6). These are used
means of a 24-bp translation initiation region (TIR) retrieved from for amplification and analyses of possible inserts. Once the gene
the GFP variant borne by pGreenTIR plasmid (27). This is an encoding the inducer-responsive TF is placed in this arrangement
unusual ribosome-binding sequence known to act as a transla- as a NcoI (overlapping the leading ATG)–PacI DNA fragment, the
tional enhancer that is expected to curb the expression noise that whole expression module becomes inserted in the SEVA frame as a
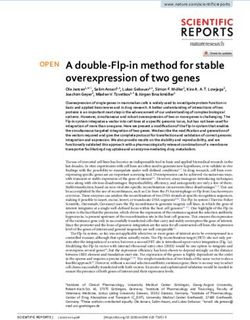
Figure 3. Transcriptional activity of AlkS/PalkB node before and after standardization as a SEVA cargo. P. putida transformants with either (A) pARalkS
(wild-type alkS) or (B) pSEVA429 crc+ → GFP (edited alkS variant ED) were grown in LB until mid-exponential phase and treated or not with 0.05% AlkS
effector DCPK. Fluorescent emission was then measured in a cytometer for the next 6 h as explained in Section 2. The right panels show the mean GFP
fluorescence signals with their respective standard deviation calculated from the populations showed in (A) and (B). The experiments were carried out
in biological triplicates.6 Synthetic Biology, 2021, Vol. 6(1), No. 0
PacI–AvrII addition (Figure 1C), shielded both upstream and down- Supplementary Figure S1. The alkSED variant was then coupled to
stream by transcriptional terminators contributed by the vector the spacer shown in Figure 1B, which was added with the wild-
structure and ready to be inserted with any gene of interest cloned type 90-bp sequence of the target PalkB promoter (see Section 2).
in the corresponding polylinker. The resulting cargo was subsequently placed in plasmid pSEVA421
(7) as a PacI–AvrII insert, resulting in expression vector pSEVA429
3.2 Reshaping the alkS and PalkB pair as an crc+ (Table 1; Supplementary Figure S2). For parameterization of
inducible expression device the activity of the thereby refactored expression device, the con-
As a case study of formatting a naturally occurring inducible pro- struct was added with the promoterless GFP tir gene of pSEVA427
moter into a standardized expression cargo, we picked the regula- (7) downstream of PalkB (Section 2) resulting in reporter plasmid
tory elements that control transcription of the alk genes of the OCT pSEVA429 crc+ → GFP. To have a reference of the wild-type expres-
plasmid of P. putida GPo1 (8, 9). The choice was motivated by the sion device borne by plasmid pOCT with native regulatory parts,
exemplary regulatory density of the native system that includes we excised the ∼6.7-kb NotI fragment of plasmid pJAMA30 (12)
transcriptional and post-transcriptional control layers checking containing alkST and a divergent PalkB promoter upstream of a GFP
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
expression of alkS (16, 30; Figure 2A). This provided an archetypal reporter (Supplementary Figure S3). This DNA was then inserted
case to inspect the impact of the simplified formatting explained into the same plasmid frame of pSEVA421 used for the edited
above on the behavior of the resulting expression module. To this devices, thereby originating control plasmid pARalkS (Table 1; note
end, we first edited the wild-type DNA sequence of the regulator to that the alkS variant in this case is the original wild-type alkSWT as
eliminate restriction sites incompatible with the SEVA rules while shown in Supplementary Figure S1). Once equivalent constructs
preserving the amino acid sequence. The resulting DNA segment with alkSWT (pARalkS) and alkSED (pSEVA429 crc+ → GFP) were con-
was then produced as a 5′ → 3′ 2668-bp NcoI–PacI DNA fragment. structed, we were able to evaluate the effect of the standardiza-
The complete list of nucleotide changes entered in the sequence tion of the architecture of the regulatory node on transcriptional
of this alkS variant (that we term alkSED for edited) is compiled in performance.
Figure 4. Effect of Crc on performance of the standardized AlkS/PalkB module. Plasmids (A) pARalkS (alkSWT ) and (B) pSEVA429crc+ → GFP (alkSED )
were placed in a crc::Gm mutant of P. putida KT2440, the transformants grown in LB and treated as indicated in the legend of Figure 3. The right panels
show the mean GFP fluorescence signals with their respective standard deviation calculated from the populations shown in (A) and (B). The
experiments were carried out in biological triplicates.A. Arce-Rodríguez et al. 7
3.3 The effect of formatting AlkS/PalkB on the Finally, a closer inspection of cells not added with DCPK revealed
performance of the expression system a degree of non-induced basal transcription (in particular in the
In the first series of experiments, we compared the behavior of the wild-type, non-edited configuration; Supplementary Figure S4)
AlkS/PalkB pair assembled with all the native regulatory systems that spontaneously increased with growth (Figure 3A). Although
borne by the pOCT plasmid (Figure 2A) and recreated in plasmid such a basal level is not high (it remains within the same order of
pARalkS versus that of the same regulatory system arranged with magnitude than the baseline expression), it has to be taken into
the composition standard of Figure 1 as implemented in pSEVA429 account when expression of toxic proteins or coupling with other
crc+ → GFP. Differences included a synthetic expression segment devices is pursued (33). These results confirmed the correct func-
for transcription and translation of the regulator and an upstream tioning of the expression device following the reassembly of its
buffer DNA sequence that was followed by the divergent AlkS tar- DNA parts. Yet, we wondered about the less predictable effects
get promoter PalkB as sketched in Figure 2B. P. putida KT2440 was of other physiological inputs that operate on the AlkS/PalkB in its
transformed with each of these two plasmids, and transformants native context.
were grown in LB medium with Sm and fluorescent readout fol-
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
lowed as explained in Section 2. The cytometry results of these 3.4 Effect of Crc on the formatted and
experiments are shown in Figure 3. Inspection of the resulting not-formatted AlkS/PalkB device
graphs revealed some features of both the native and the format- Ideally, for engineering-reliable genetic devices, regulatory parts
ted system that are worth considering for handling the expression should deliver their function in a context-independent manner
device. First, whether formatted or not, both PalkB systems had a (34). Such a context includes not only genomic locations (35) and
comparable dynamic range (i.e. absolute promoter output; 31) and availability of resources (36) but also physiological signals (37, 38)
a similar quasi-linear induction kinetics (differences were ∼20%). that orchestrate the induction hierarchy. One of these is catabo-
Second, both devices displayed an increasingly sharp unimodal lite repression (30), which in the case of P. putida operates through
expression pattern (32) along the induction period with low noise a complex interplay between the proteins Crc and Hfq with small
and limited cell-to-cell variation at the time of full induction. RNAs to inhibit translation of messenger RNAs (mRNAs) of target
Figure 5. Behavior of an alkS gene variant devoid of the Crc/Hfq-binding site to its mRNA. Plasmid pSEVA429 → GFP (with the AlkS/PalkB device bearing
the regulator gene devoid of the Crc-binding site in its mRNA) was transformed into crc+ (A) and crc− (B) strains of P. putida. Transformants were grown
in LB and treated as indicated in the legend of Figure 3. The right panels of the figure show the mean GFP fluorescence signals with their respective
standard deviation that were calculated from the populations shown in (A) and (B). The experiments were carried out in biological triplicates.8 Synthetic Biology, 2021, Vol. 6(1), No. 0
genes (16). AlkS is subject to such post-transcriptional regulation
when placed in P. putida but not in E. coli (39). In order to cali-
brate the effect of such a control layer and whether it was kept or
not in the standardized construct, we run the experiments shown
in Figure 4. To this end, pARalkS and pSEVA429 crc+ → GFP were
placed in an isogenic crc::Gm mutant of P. putida (Table 1) known
to be blind to catabolite repression caused by many of the compo-
nents of LB medium (40, 41). As shown in Figure 4, the overall
behavior of the standardized and non-standardized expression
devices remained very similar in dynamic range and inducibility,
although their expression kinetics was slower as compared to the
data shown in Figure 3.
The data above were welcome results, as refactoring of an
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
existing regulatory node with a different architecture often results
in devices that perform significantly worse than the naturally
occurring setup (42, 43). But the question still remained of
whether we could erase altogether the effect of crc on the per-
formance of the AlkS/PalkB module, not by moving to a different
genetic background but by rewriting the DNA sequence of the
regulator. To address this, an additional alkS derivative was syn-
thesized in which the Hfq/Crc-binding site of the corresponding Figure 6. Influence of different inducer concentrations in P. putida
mRNA was edited as described in Section 2 and Supplementary populations bearing the AlkS/PalkB module with the regulator gene with
or without the Crc-binding site. Plasmids pSEVA429 crc+ → luxCDABE
Figure S1. These changes replace the original codons by synonym
(alkSED , with Crc-binding site) and pSEVA429 → luxCDABE (alkSCR ,
triplets in a way that keeps the amino acid sequence of the AlkS without Crc site) were transformed into P. putida KT2440, grown in LB
protein but breaks the target site for Hfq/Crc within the cog- until mid-exponential phase and added with the DCPK concentrations
nate mRNA. As before, the resulting DNA (that bears the variant indicated in each case. Luminescent emission was then recorded after
4 h as a proxy of transcriptional activity as explained in Section 2. (A)
hereafter called alkSCR for Crc-free) was formatted as a NcoI–PacI
Sketch, not to scale, of functional segments in reporter plasmids. (B)
fragment and coupled to the spacer shown in Figure 1B and the Readout of reporter constructs borne by P. putida. Data shown for three
PalkB promoter. The resulting cargo was subsequently placed in technical replicates.
the frame of plasmid pSEVA421 as PacI–AvrII insert, resulting in
expression vector pSEVA429 (Supplementary Figure S2). For the
sake of comparing its performance, a promoterless GFP gene iden- the Crc/Hfq-based system of catabolic repression that is typical
tical to that of pARalkS and pSEVA429 crc+ → GFP was added of the Pseudomonads (34), while the second is a close—but by no
to pSEVA429, thereby generating pSEVA429 → GFP. This alkSCR - means identical—relative of P. putida. For the experiments, E. coli
containing plasmid was then placed in isogenic crc+ and crc− CC118 and P. aeruginosa PAO1 were transformed with pSEVA429
strains of P. putida, and the readout of the fluorescent reporters → GFP bearing the edited the alkSCR/ PalkB device described above,
followed in LB medium with or without DCPK induction as before. grown and induced with DCPK as before. The results are shown in
As shown in Figure 5, removal of the crc site from the alkS sequence Figure 7. While the gross inducibility pattern was kept in either
had only a moderate effect on the performance of the expres- case, the parameters changed significantly, especially when E.
sion device, as the induction patterns were quite similar when coli was used as the host of the construct (Figure 7A). In this
pSEVA429 → GFP was placed in isogenic P. putida strains with or case, the inducibility and the dynamic range of the promoter
without the factor. were worse than P. putida with the same plasmid (Figure 5A), and
To gain some insight into this apparently minor influence of the expression pattern was noisy—as compared to the basically
removing the Crc site in alkS, we constructed additional deriva- monomodal display of the same device in the original host. In con-
tives of pSEVA429 crc+ and pSEVA429 with a promoterless lux- trast, when the host of pSEVA429 GFP was P. aeruginosa (Figure 7B)
CDABE operon. This luminescent reporter is considerably more and despite a conspicuous increase of phenotypic diversity, the
sensitive than GFP (44) and therefore a better proxy of transcrip- expression module behaved within the same parameter set than
tional output from PalkB at a population level. The new construct that observed using P. putida as the host (Figure 5A). This was not a
was placed in P. putida KT2440 and the cognate transformants surprise given the physiological and regulatory similarity between
grown in LB with different inducer concentrations. In this case the two Pseudomonas strains used.
(Figure 6), the overall effect of removing the Crc site from the
alkS mRNA became more evident. As shown in Figure 6, the con-
struct with alkSCR produced luminescence levels approximately 4. Conclusions
twice as high those as the same with alkSED . This accredited the In this work, we have used the inducer-dependent and AlkS-
role of Crc on alkS expression and its improvement with the edited mediated activation of the PalkB promoter of the pOCT plasmid as
gene, although we cannot rule out that factors other than the an example of the roadmap that could be followed for reshaping
transcriptional regulator can also influence physiological control. a naturally occurring regulatory node into a standardized device
for engineering heterologous expression (45). As with any stan-
3.5 Interoperability of the alkSCR/ PalkB device in dard, there is an arbitrary but still reasonable and scientifically
other bacteria justifiable choice of a given composition rule (46). The one we
Once the expression module engineered in pSEVA429 was char- propose in this work is summarized in Figure 1 and explained in
acterized in P. putida, we next examined its performance in E. coli detail above. As is also the case of other standards, this particular
and P. aeruginosa (Table 1). Note that the first host lacks altogether choice will certainly limit flexibility but will foster interoperability,A. Arce-Rodríguez et al. 9
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
Figure 7. Behavior of the alkSCR gene variant devoid of the Crc/Hfq-binding site in other bacterial species. Plasmid pSEVA429 → GFP was transformed
into (A) E. coli DH5α or (B) P. aeruginosa PAO1. Transformants were grown in LB and treated as indicated in the legend of Figure 3. The right panels of
the figure show the mean GFP fluorescence signals with their respective standard deviation that were calculated from the populations shown in (A)
and (B). Each experiment was carried out in biological triplicates.
parameterization and comparative metrology (47, 48). The work performance with other expression modules that follow the same
above exemplifies how the same device, still after formatting, may arrangement. We ultimately expect such standardization to ease
go through successive, improved versions of the same functional the engineering of complex systems and encourage other genetic
DNA segments even if a prefixed arrangement is kept constant. In tool developers to follow suit.
the cases examined above, we can consider plasmids pARalkS and
pSEVA429 crc+ as beta versions of what we propose to be an stan-
dardized AlkS/PalkB expression device apt for inclusion as a cargo
Supplementary data
in the SEVA collection: pSEVA429 (Supplementary Figure S2). A Supplementary data are available at SYNBIO Online.
comparative summary of the more relevant properties of the key
constructs with the alkSCR/ PalkB module used in this work in the
hosts of interest is shown in Table 2. Despite the considerable edits
Data availability
of the regulatory parts involved and their relative position within All materials described in this article are available upon reason-
the same DNA segment, it is noteworthy that the basic proper- able request.
ties were kept in the standardized version. Note however that—as
shown above—there is still room for improvement, and it is likely
that other versions will follow, an issue that is contemplated in the
Funding
updated nomenclature of the SEVA collection (7). For instance, the SETH (RTI2018-095584-B-C42) (MINECO/FEDER); SyCoLiM (ERA-
system could be refactored to make it more digital (e.g. lowering COBIOTECH 2018-PCI2019-111859-2) Projects of the Spanish
its basal expression (33)), making it more independent of physio- Ministry of Science and Innovation; MADONNA (H2020-FET-
logical control. But, in the meantime, pSEVA429 is an altogether OPEN-RIA-2017-1-766975); BioRoboost (H2020-NMBP-BIO-CSA-
standardized off-the-shelf expression vector with a large number 2018-820699); SynBio4Flav (H2020-NMBP-TR-IND/H2020-NMBP-
of benefits, including the possibility of comparing faithfully its BIO-2018-814650); MIX-UP (MIX-UP H2020-BIO-CN-2019-870294)10 Synthetic Biology, 2021, Vol. 6(1), No. 0
Contracts of the European Union; InGEMICS-CM (S2017/BMD- 13. Makart,S., Heinemann,M. and Panke,S. (2007) Characterization
3691) Project of the Comunidad de Madrid-European Structural of the AlkS/PalkB-expression system as an efficient tool for the
and Investment Funds (FSE, FECER). production of recombinant proteins in Escherichia coli fed-batch
fermentations. Biotechnol. Bioeng., 96, 326–336.
14. Reed,B., Blazeck,J. and Alper,H. (2012) Evolution of an alkane-
Acknowledgments inducible biosensor for increased responsiveness to short-chain
Authors are indebted to Fernando Rojo and Renata Moreno for alkanes. J. Bacteriol., 158, 75–79.
strains and valuable materials. 15. Bahls,M.O., Kardashliev,T. and Panke,S. (2017) Novel sensors for
engineering microbiology. In: Lee SY (ed). Consequences of Microbial
Interactions with Hydrocarbons, Oils, and Lipids: Production of Fuels and
Conflict of interest statement. None declared.
Chemicals. Springer, New York, pp. 331–357.
16. Hernández-Arranz,S., Sánchez-Hevia,D., Rojo,F. and Moreno,R.
References (2016) Effect of Crc and Hfq proteins on the transcription, pro-
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
1. Terpe,K. (2006) Overview of bacterial expression systems for het- cessing, and stability of the Pseudomonas putida CrcZ sRNA. RNA,
erologous protein production: from molecular and biochemical 22, 1902–1917.
fundamentals to commercial systems. Appl. Microbiol. Biotech., 72, 17. Dinamarca,M.A., Ruiz-Manzano,A. and Rojo,F. (2002) Inactivation
211–222. of cytochrome o ubiquinol oxidase relieves catabolic repression
2. Browning,D.F., Godfrey,R.E., Richards,K.L., Robinson,C. and of the Pseudomonas putida GPo1 alkane degradation pathway. J.
Busby,S.J.W. (2019) Exploitation of the Escherichia coli lac operon Bacteriol., 184, 3785–3793.
promoter for controlled recombinant protein production. Biochem. 18. Sambrook,J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning:
Soc. Trans., 47, 755–763. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold
3. Fernandez-López,R., Ruiz,R., de la Cruz,F. and Moncalián,G. (2015) Spring Harbor, NY.
Transcription factor-based biosensors enlightened by the analyte. 19. de Lorenzo,V. and Timmis,K.N. (1994) Analysis and construction of
Front Microbiol., 6, 648. stable phenotypes in gram-negative bacteria with Tn5- and Tn10-
4. Marschall,L., Sagmeister,P. and Herwig,C. (2017) Tunable recom- derived minitransposons. Methods Enzymol., 235, 386–405.
binant protein expression in E. coli: promoter systems and genetic 20. Choi,K.H., Kumar,A. and Schweizer,H.P. (2006) A 10-min method
constraints. Appl. Microbiol. Biotech., 101, 501–512. for preparation of highly electrocompetent Pseudomonas aerugi-
5. Martínez-García,E. and de Lorenzo,V. (2017) Molecular tools and nosa cells: application for DNA fragment transfer between chro-
emerging strategies for deep genetic/genomic refactoring of Pseu- mosomes and plasmid transformation. J. Microbiol. Methods, 64,
domonas. Curr. Opin. Biotechnol., 47, 120–132. 391–397.
6. Silva-Rocha,R., Martinez-Garcia,E., Calles,B., Chavarria,M., 21. Yuste,L., Canosa,I. and Rojo,F. (1998) Carbon-source-dependent
Arce-Rodriguez,A., de las Heras,A., Paez-Espino,A.D., expression of the PalkB promoter from the Pseudomonas oleovorans
Durante-Rodriguez,G., Kim,J., Nikel,P.I. et al. (2013) The Standard alkane degradation pathway. J. Bacteriol., 180, 5218–5226.
European Vector Architecture (SEVA): a coherent platform for the 22. Horton,R.M., Hunt,H.D., Ho,S.N., Pullen,J.K. and Pease,L.R. (1989)
analysis and deployment of complex prokaryotic phenotypes. Engineering hybrid genes without the use of restriction enzymes:
Nucleic Acids Res., 41, D666–D675. gene splicing by overlap extension. Gene, 77, 61–68.
7. Martínez-García,E., Goñi-Moreno,A., Bartley,B., McLaughlin,J., 23. Nielsen,A.A.K., Der,B.S., Shin,J., Vaidyanathan,P., Paralanov,V.,
Sánchez-Sampedro,L., Del Pozo,H.P., Hernández,C.P., Strychalski,E.A., Ross,D., Densmore,D. and Voigt,C.A. (2016)
Marletta,A.S., De Lucrezia,D., Sánchez-Fernández,G. et al. (2020) Genetic circuit design automation. Science, 352, aac7341.
SEVA 3.0: an update of the Standard European Vector Architec- 24. Thompson,M.G., Pearson,A.N., Barajas,J.F., Cruz-Morales,P.,
ture for enabling portability of genetic constructs among diverse Sedaghatian,N., Costello,Z., Garber,M.E., Incha,M.R.,
bacterial hosts. Nucleic Acids Res., 48, 3395. Valencia,L.E., Baidoo,E.E.K. et al. (2020) Identification,
8. Rojo,F. (2009) Degradation of alkanes by bacteria. Environ. Micro- characterization, and application of a highly sensitive lactam
biol., 11, 2477–2490. biosensor from Pseudomonas putida. ACS Synth. Biol., 9, 53–62.
9. van Beilen,J.B., Panke,S., Lucchini,S., Franchini,A.G., 25. Alon,U. (2019) An Introduction to Systems Biology: Design Principles of
Röthlisberger,M. and Witholt,B. (2001) Analysis of Pseudomonas Biological Circuits. CRC Press, Boca Raton, FL.
putida alkane-degradation gene clusters and flanking insertion 26. Martínez-García,E., Calles,B., Arévalo-Rodríguez,M. and de
sequences: evolution and regulation of the alk genes. Microbiology, Lorenzo,V. (2011) pBAM1: an all-synthetic genetic tool for anal-
147, 1621–1630. ysis and construction of complex bacterial phenotypes. BMC
10. Canosa,I., Sánchez-Romero,J.M., Yuste,L. and Rojo,F. (2000) A Microbiol., 11, 38.
positive feedback mechanism controls expression of AlkS, the 27. Miller,W.G. and Lindow,S.E. (1997) An improved GFP cloning cas-
transcriptional regulator of the Pseudomonas oleovorans alkane sette designed for prokaryotic transcriptional fusions. Gene, 191,
degradation pathway. Mol. Microbiol., 35, 791–799. 149–153.
11. Sticher,P., Jaspers,M.C., Stemmler,K., Harms,H., Zehnder,A.J. and 28. Amann,E., Ochs,B. and Abel,K.-J. (1988) Tightly regulated tac pro-
van der Meer,J.R. (1997) Development and characterization of a moter vectors useful for the expression of unfused and fused
whole-cell bioluminescent sensor for bioavailable middle-chain proteins in Escherichia coli. Gene, 69, 301–315.
alkanes in contaminated groundwater samples. Appl. Environ. 29. Wu,H.-Y., Shyy,S., Wang,J.C. and Liu,L.F. (1988) Transcription gen-
Microbiol., 63, 4053. erates positively and negatively supercoiled domains in the tem-
12. Jaspers,M.C., Meier,C., Zehnder,A.J., Harms,H. and plate. Cell, 53, 433–440.
van der Meer,J.R. (2001) Measuring mass transfer processes 30. Rojo,F. (2010) Carbon catabolite repression in Pseudomonas: opti-
of octane with the help of an alkS alkB:: gfp-tagged Escherichia coli. mizing metabolic versatility and interactions with the environ-
Env. Microbiol., 3, 512–524. ment. FEMS Microbiol. Revs., 34, 658–684.A. Arce-Rodríguez et al. 11
31. Snoek,T., Chaberski,E.K., Ambri,F., Kol,S., Bjørn,S.P., Pang,B., 43. Temme,K., Zhao,D. and Voigt,C.A. (2012) Refactoring the nitrogen
Barajas,J.F., Welner,D.H., Jensen,M.K. and Keasling,J.D. (2020) fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci.
Evolution-guided engineering of small-molecule biosensors. USA, 109, 7085–7090.
Nucleic Acids Res., 48, e3. 44. Sagi,E., Hever,N., Rosen,R., Bartolome,A., Premkumar,J., Ulber,R.,
32. Kærn,M., Elston,T.C., Blake,W.J. and Collins,J.J. (2005) Stochasticity Lev,O., Scheper,T. and Belkin,S. (2003) Fluorescence and biolu-
in gene expression: from theories to phenotypes. Nat. Revs. Genet., minescence reporter functions in genetically modified bacterial
6, 451–464. sensor strains. Sens. Actuators, 90, 2–8.
33. Calles,B., Goñi-Moreno,Á. and de Lorenzo,V. (2019) Digitalizing 45. Martínez-García,E., Benedetti,I., Hueso,A. and de Lorenzo,V.
heterologous gene expression in Gram-negative bacteria with a (2015) Mining environmental plasmids for synthetic biology parts
portable ON/OFF module. Mol. Syst. Biol., 15, e8777. and devices. Microbiol. Spectr., 3, Plas-0033-2014.
34. Costello,A. and Badran,A.H. (2021) Synthetic biological circuits 46. Porcar,M., Danchin,A. and de Lorenzo,V. (2015) Confidence, toler-
within an orthogonal central dogma. Trends Biotechnol., 39, ance, and allowance in biological engineering: the nuts and bolts
59–71. of living things. Bioessays, 37, 95–102.
Downloaded from https://academic.oup.com/synbio/article/6/1/ysab030/6371770 by guest on 28 December 2021
35. Bryant,J.A., Sellars,L.E., Busby,S.J.W. and Lee,D.J. (2014) Chromo- 47. de Lorenzo,V. and Schmidt,M. (2018) Biological standards for the
some position effects on gene expression in Escherichia coli K-12. Knowledge-Based BioEconomy: what is at stake. New Biotechnol.,
Nucleic Acids Res., 42, 11383–11392. 40, 170–180.
36. Borkowski,O., Ceroni,F., Stan,G.-B. and Ellis,T. (2016) Overloaded 48. Beal,J., Goñi-Moreno,A., Myers,C., Hecht,A., de Vicente,M.D.C.,
and stressed: whole-cell considerations for bacterial synthetic Parco,M., Schmidt,M., Timmis,K., Baldwin,G., Friedrichs,S. et al.
biology. Curr. Op. Microbiol., 33, 123–130. (2020) The long journey towards standards for engineering biosys-
37. Cases,I. and de Lorenzo,V. (2005) Promoters in the environment: tems: are the molecular biology and the biotech communities
transcriptional regulation in its natural context. Nat. Revs. Micro- ready to standardise? EMBO Rep., 21, e50521.
biol., 3, 105–118. 49. Manoil,C. and Beckwith,J. (1985) TnphoA: a transposon probe for
38. Shingler,V. (2011) Signal sensory systems that impact σ54- protein export signals. Proc. Natl. Acad. Sci. USA, 82, 8129–8133.
dependent transcription. FEMS Microbiol. Revs., 35, 425–440. 50. Hanahan,D. and Meselson,M. (1983) Plasmid screening at high
39. Yuste,L. and Rojo,F. (2001) Role of the crc gene in catabolic repres- colony density. Methods Enzymol., 100, 333–342.
sion of the Pseudomonas putida GPo1 alkane degradation pathway. 51. Nelson,K.E., Weinel,C., Paulsen,I.T., Dodson,R.J., Hilbert,H.,
J. Bacteriol., 183, 6197–6206. Martins dos Santos,V.A., Fouts,D.E., Gill,S.R., Pop,M., Holmes,M.
40. Molina,L., La Rosa,R., Nogales,J. and Rojo,F. (2019) Influence of the et al. (2002) Complete genome sequence and comparative analy-
Crc global regulator on substrate uptake rates and the distribu- sis of the metabolically versatile Pseudomonas putida KT2440. Env.
tion of metabolic fluxes in Pseudomonas putida KT2440 growing in Microbiol., 4, 799–808.
a complete medium. Environ. Microbiol., 21, 4446–4459. 52. Holloway,B.W. (1955) Genetic recombination in Pseudomonas aerug-
41. Molina,L., Rosa,R.L., Nogales,J. and Rojo,F. (2019) Pseudomonas inosa. J. Gen. Microbiol., 13, 572–581.
putida KT2440 metabolism undergoes sequential modifications 53. Kessler,B., de Lorenzo,V. and Timmis,K.N. (1992) A general system
during exponential growth in a complete medium as compounds to integrate lacZ f usions into the chromosomes of gram-negative
are gradually consumed. Environ. Microbiol., 21, 2375–2390. eubacteria: regulation of the Pm promoter of the TOL plasmid
42. Chan,L.Y., Kosuri,S. and Endy,D. (2005) Refactoring bacteriophage studied with all controlling elements in monocopy. Mol. Gen.
T7. Mol. Sys. Biol., 1, 2005.0018. Genet., 233, 293–301.You can also read