Cloning of the Human Keratin 18 Gene and Its Expression in Nonepithelial Mouse Cells
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
MOLECULAR AND CELLULAR BIOLOGY, Apr. 1988, p. 1540-1550 Vol. 8, No. 4
0270-7306/88/041540-11$02.00/0
Copyright X 1988, American Society for Microbiology
Cloning of the Human Keratin 18 Gene and Its Expression in
Nonepithelial Mouse Cells
DAVID A. KULESH AND ROBERT G. OSHIMA*
Cancer Research Center, La Jolla Cancer Research Foundation, 10901 North Torrey Pines Road, La Jolla,
California 92037
Received 1 December 1987/Accepted 1 January 1988
Human keratin 18 (K18) and the homologous mouse protein, Endo B, are intermediate filament subunits of
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
the type I keratin class. Both are expressed in many simple epithelial cell types including trophoblasts, the first
differentiated cell type to appear during mouse embryogenesis. The K18 gene was identified and cloned from
among the 15 to 20 similar sequences identified within the human genome. The identity of the cloned gene was
confirmed by comparing the sequence of the first two exons to the K18 cDNA sequence and transfecting the gene
into various murine cell lines and verifying the encoded protein as K18 by immunoprecipitation and partial
peptide mapping. The transfected K18 gene was expressed in mouse HR9 parietal endodermal cells and mouse
fibroblasts even though the fibroblasts fail to express endogenous Endo B. Si nuclease protection analysis
indicated that mRNA synthesized from the transfected K18 gene is initiated at the same position as authentic
K18 mRNA found in both BeWo trophoblastoma cells and HeLa cells. Pulse-chase experiments indicated that
the human K18 protein is stable in murine parietal endodermal cells (HR9) which express EndoA, a
complementary mouse type II keratin. Surprisingly, however, K18 was degraded when synthesized in cells
which lack a type II keratin. This turnover of K18 may be an important mechanism by which epithelial cells
maintain equal molar amounts of both type I and II keratins. In addition, the levels of the endogenous type I
Endo B in parietal endodermal cells were compensatingly down regulated in the presence of the K18 protein,
while the levels of the endogenous type II Endo A were not affected in any of the transfected cell lines.
Intermediate filament (IF) genes are expressed in nearly proteins expressed during the development of the mouse (13,
all mammalian cells, resulting in polypeptide subunits which 14, 31, 55). These IF proteins are located in the trophoblast
polymerize to form relatively insoluble filaments of approx- layer of the blastocyst-stage embryo (13, 55) and are ulti-
imately 10 nm diameter. These proteins are categorized into mately found in a limited number of adult differentiated cell
at least five groups: vimentin, desmin, glial fibrillary acidic types.
proteins, three neurofilament proteins, and cytokeratins (69) In vitro studies of these developmentally regulated kera-
in addition to a sixth class composed of the nuclear lamins tins utilize mouse embryonal carcinoma (EC) cells (13, 53)
(24, 44). Vimentin is characteristically expressed by cells of and human EC cells (1, 2). F9, a mouse EC stem cell line,
mesenchymal origin, although in vitro culturing of cells from does not express Endo A or Endo B until the cells are
diverse vertebrates commonly induces vimentin synthesis induced to differentiate to extraembryonic endoderm (53,
(25). Desmin is primarily expressed in muscle cells, although 54). Human embryonal carcinoma cells, however, already
a transfected desmin gene can be expressed in nonmuscle express K18 and its coexpressed type II partner, K8 (18, 19;
cells (61). Glial fibrillary acidic proteins are expressed ex- unpublished data). Consideration of the tissue-specific
clusively in certain glial cell types, while neurofilament expression of K18 and Endo B in adult tissues, the early
protein expression appears to be limited to neuronal cells developmental expression of the two genes, and the appar-
(35). The largest class of IF proteins, the cytokeratins, have ent coordinate regulation of the levels of the type I and type
been further subdivided into either type I (acidic) or type II II keratin subunits within cells raises several questions
(basic) keratins (23, 70) with at least one of each type which could be investigated once the appropriate genes are
necessary for filament formation (68). Particular type I and isolated. Here we report the isolation and expression of the
type II keratins are expressed as preferential pairs that are gene coding for K18. The K18 gene could be expressed in
usually found in equal proportion within cells. However, mouse fibroblasts which do not express the homologous
filaments can be formed in vitro from type I and type II Endo B gene. However, in the absence of a type II keratin
subunits which are not normally coexpressed in vivo (29). subunit, the protein was turned over relatively rapidly. In
The genes which code for several of the type I and II contrast, expression of the K18 gene in mouse parietal
epidermal keratins have been isolated and characterized (3, endodermal cells that contain a compatible type II keratin
32, 34, 41, 42, 59, 62). The overall structure of most IF genes resulted in expression of a stable K18 protein and down
is highly conserved with respect to their intron positions regulation of the homologous murine gene product, Endo B.
(41), the exception being a neurofilament protein gene (36).
The expression of IF proteins during early development MATERIALS AND METHODS
has been investigated primarily in the mouse. Endo A and Cells and media. The F9.22 mouse embryonal carcinoma
Endo B, the mouse forms of human keratin 8 (K8) and (EC) cell line (7, 52), HR9 mouse parietal endodermal cell
keratin 18 (K18) (49), respectively, appear to be the first IF line (15), and STO mouse fibroblast line (43, 53) have been
described previously. L cells were a thymidine kinase-
*
Corresponding author. deficient derivative obtained from W. Rashke (La Jolla
1540VOL. 8, 1988 CLONING AND EXPRESSION OF HUMAN KERATIN 18 GENE 1541
Cancer Research Foundation). All cells were maintained in by plaque hybridization (6). After three rounds of purifica-
Dulbecco modified Eagle medium supplemented with pyru- tion and screening, 10 positive Charon 30 plaques were
vate (110 mg/ml), 0.04% glutamine, 10% (vol/vol) fetal bo- isolated for further analysis. The 9.9-kb fragment of one
vine serum, and gentamicin sulfate (30 ,ug/ml) as previously phage isolate (recombinant Charon 30 clone 18.23) which
described (53, 54). hybridized to both the 5' and 3' cDNA probes was subcloned
Strains. Escherichia coli VCS257 (a derivative of DP50 into the Hindlll site of pGEM-1, yielding plasmids pGC1853
SupF selected for lambda plating efficiencies; Stratagene, and pGC1835.
San Diego, Calif.) and E. coli K802 (39) were used as hosts DNA transfections. Isolated plasmid DNAs were transfec-
for the cloning vector Charon 30. E. coli DH-1 (39) was used ted into HR9, L, F9.22, and STO mouse cells by the calcium
as a host for plasmids. E. coli JM101 (47) was used as the phosphate method (38). Transfections with pGC1853 were
host for the propagation of M13 bacteriophage. performed with additional pSV2Neo (66) at a 10:1 (wt/wt)
Plasmid constructions. Restriction fragments were purified ratio to provide a selectable marker for integrated DNA.
from agarose gels by either electroelution or adsorption to Immunoprecipitation and partial peptide mapping. Meta-
powdered glass (73) (BiolOl, San Diego, Calif.). Plasmids bolic labeling of cells with [35S]methionine (100 ,uCi/ml),
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
pK187 and pK189 consist of the full-length K18 cDNA lysate preparation, immunoprecipitation, and gel electropho-
cloned into the EcoRI multiple cloning site of pGEM-1 in resis were performed as previously described (54) except
two different orientations (56). pK187 has the 5' end of the that soybean trypsin inhibitor was added at a final concen-
K18 cDNA adjacent to the T7 RNA polymerase promoter, tration of 100 ,ug/ml to the final wash of the cells and to the
while pK189 has the 3' end of the cDNA adjacent to the T7 lysate. Antisera to Endo A and Endo B were used to
promoter. Plasmids pGC1853 and pGC1835 consist of the precipitate Endo A and K8 or Endo B and K18, respectively
entire 9.9-kilobase (kb) HindIII human DNA fragment that (53). All immunoprecipitations were performed in antibody
contains the K18 gene. pGC1853 has the 5' end of the K18 excess with the same amount of trichloroacetic acid-insol-
gene proximal to the T7 promoter of pGEM-1, while uble radioactive lysate. Film exposures were quantitated by
pGC1835 has the insert in the opposite orientation. Plasmid densitometry with the Hoeffer GS-360 scanning densitom-
pGC39 was constructed by inserting the 3.9-kb BamHI eter system and appropriately exposed films.
fragment (nucleotides [nt] -2526 to 1379) from pGC1853 into Partial peptide mapping by limited proteolysis in sodium
the BamHI site of pGEM-1. The 5' BamHI site of this dodecyl sulfate (SDS) was performed as previously de-
fragment was derived from the pGEM-1 polylinker. The scribed (16). Briefly, 107 cpm of cell lysate were immuno-
plasmid K18pONeo contains the 5'-flanking region, exon 1, precipitated with 10 VI of Endo B antiserum. Trypsin (0.6
and intron 1 of the K18 gene upstream of the Tn5 neo gene p.g/ml, final concentration) was added to the eluted precipi-
coding region and the simian virus 40 (SV40) small-t-antigen tate and incubated at 37°C for 15 min. The reaction was
splice and polyadenylation signals. It was constructed by stopped by the addition of mercaptoethanol and SDS and
inserting neo and SV40 sequences derived from pSV2Neo heating to 100°C. Each sample was resolved on an 18%
(66) by digestion with BglII and BamHI into the unique BglII acrylamide-SDS gel (53) and processed for fluorography
site of pGC39. The neo gene fragment contains translational (11).
stop codons in all three reading frames upstream or imme- DNA sequence analysis. The 3.9-kb 5' end of the K18 gene
diately downstream of the AUG initiating codon. Thus, the was excised from pGC1853 by digestion with BamHI and
expected mRNA from this construction would code for one subcloned into M13mpl8. Deletions were generated with
spliced mRNA that could be translated into a truncated K18 exonuclease III by the method of Henikoff (30) after diges-
polypeptide and the phosphotransferase enzyme of the neo tion with XhoI and SphI. For determination of the sequence
gene (57, 58). of the opposite strand, the 3.9-kb fragment was isolated by
Isolation of K18 genomic clones. Eucaryotic DNA was digestion with both BamHI and HindIII and subcloned into
isolated from cells grown in culture as previously described M13mpl9. Deletions were generated by the same method
(71). K18 cDNA was labeled with [a-32P]dCTP (>600 after digestion with KpnI and BamHI. All sequencing was
Ci/mmol; New England Nuclear Corp., Boston, Mass.) by performed by the method of Sanger (64) modified to use
nick translation (45). The 5' K18 riboprobes were prepared 7-deaza-2'-dGTP (5, 48) and deoxyadenosine 5'-[a-355]thio-
from plasmid pK187 that had been digested with either NciI triphosphate (8). The DNA sequence data were compiled by
or BstNI. NciI cuts at nt 43 within the 5' noncoding region, the GEL program of the BIONET computer system (Intel-
while BstNI cuts at nt 106 within the 5' coding region. The 3' liGenetics, Mountain View, Calif.). The final sequence was
K18 riboprobe was prepared from RsaI-digested pK189. assembled from 65 separate sequencing reactions, 22 isolates
RsaI cuts at nt 1363 in the 3' noncoding region of the K18 for one strand and 18 for the opposite strand.
cDNA. The riboprobes were synthesized with [a-32P]GTP Si analysis of RNA. RNA was isolated by the guanidinium
(>600 Ci/mmol; New England Nuclear Corp.) by T7 RNA isothiocyanate method (39). DNA probes for S1 nuclease
polymerase (46) and purified on a 3.5% polyacrylamide-8 M analysis were generated from M13 derivatives by primed
urea gel. DNA synthesis (21, 72) utilizing [32P]dATP. The probe for
HindIlI-digested HeLa cell DNA was size fractionated on K18 was derived from M13mpl9K18Sl, one of the exonu-
an agarose gel, and the region corresponding to 9 to 10 kb clease III deletions, by digesting the Klenow product with
was recovered by electroelution in several fractions. These XhoI. The probe is 471 bases long with a 240-base-pair (bp)
fractions were screened by Southern analysis with both the overlap with exon 1 of the K18 gene.
5' and 3' K18 RNA probes, and the fraction containing the The probe for Endo B RNA was prepared from a fragment
most intense hybridizing signal was cloned into the HindIll of the 5' end of the gene for Endo B (R. G. Oshima, K.
site of Charon 30 (63) by standard methods (39). The ligation Trevor, L. Shevinsky, 0. A. Ryder, and G. Cecena, submit-
products were packaged with a commercial packaging ex- ted). A 750-bp fragment generated by digestion with SstI and
tract (Gigapack Plus; Stratagene). The packaged phage were EcoRV was subcloned into M13mpl8 which had been di-
plated on E. coli VCS257 cells without prior amplification of gested with SstI and HinclI to generate M13mpl8BS1. The
the library. A total of 5 x 106 recombinants were screened sequencing primer extension product was restricted with1542 KULESH AND OSHIMA MOL. CELL. BIOL.
A B C
=- =: z= _
E oO co E o'a co E o' 0o
X C)C E
X UJ I CO Z 3c 0
toW
.C E
co
3
I
0 u 5_ E 3
mw sX0
-23.1 'o- *-23.1 W f* -23.1
%W I ".,
a WI
-9.4 %0 _0 -9.4
W.*' SV,#
ov -93.4
-6.6 * -6.6 _* -e.6
V _
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
-4.4 0 0 -44 -0 -4.4
0 0 6
o
ka
-2.3 -2.3 _ -2.3
-2.0 a _* -2.0 _* -2.0
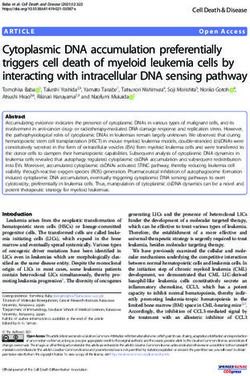
FIG. 1. Southern analysis of human K18 genes. HeLa cell DNA (10 ,ug) was digested with the indicated restriction enzyme, separated by
agarose gel electrophoresis, transferred to nitrocellulose, and hybridized to radioactive RNA probes complementary to either the 5' or 3' end
of the K18 cDNA. (A) Intensifier screen-enhanced autoradiographic image of a filter which was hybridized with the 106-nt 5' riboprobe
generated from BstNI-digested pK187 plasmid by T7 RNA polymerase. The filter was washed at 55°C in 0.1 x SSC (1x SSC is 0.15 M NaCl
plus 0.015 M sodium citrate)-0.15% SDS. (B) Same filter as in panel A after an additional wash at 70°C for 1 h. (C) Fragments which hybridized
to the 80-nt 3' riboprobe synthesized from RsaI-digested pK189 plasmid. The filter was washed at 55°C for 1 h. Lane MW, Radioactive size
markers indicated in kilobases. The asterisk (*) indicates the size-selected genomic DNA cloned into Charon 30 bacteriophage.
EcoRI, which cleaves in the polylinker approximately 513 Only a few hybridizing bands remained. These appeared to
bases upstream of the transcriptional start site of the Endo B be likely candidates to contain the coding K18 gene. Figure
cDNA, generating a probe of about 780 nucleotides with a 1C shows the result of probing the same HeLa DNA digests
195-bp overlap with exon 1 of the Endo B gene. with the K18 3' probe and washing at 55°C. One strongly
The labeled single-stranded probes were isolated by elec- hybridizing 9- to 10-kb HindlIl fragment (designated by the
trophoresis through a 1% low-melting-point agarose-alkaline asterisk) was detected with both the 5' and 3' probes,
denaturing gel and recovered by heating the appropriate gel suggesting a single DNA fragment that may contain the
slices to 65°C in 10 mM Tis-1 mM EDTA (pH 8), followed entire coding K18 gene.
by phenol, phenol-chloroform, and chloroform extractions HindIll-digested DNA of 9 to 10 kb, for which hybridiza-
and ethanol precipitation. Hybridization to the RNA was in tion with the 5' cDNA probe was confirmed in a separate
85% formamide-25 mM Tris hydrochloride (pH 7.4)-450 mM experiment, was cloned into Charon 30 bacteriophage. Ten
NaCl-10 mM EDTA-0.4% SDS at 50°C overnight as de- positive plaques were isolated for further analysis. One
scribed previously (21). The RNA-DNA hybrids were isolate which hybridized strongly to both the 5' and 3' cDNA
treated with 50 U of Si nuclease for 1 h at 37°C followed by riboprobes was analyzed further. A restriction map of the
phenol-chloroform extraction, ethanol precipitation, and insert of this isolate is compared with the known restriction
electrophoresis on a 6% polyacrylamide-8 M urea gel. sites of the K18 cDNA in Fig. 2.
The KpnI site at nt 4000 of the gene may represent an area
RESULTS close to the 3' end of the gene because the corresponding site
Isolation of K18 gene. Previously, we identified 15 to 20 in the cDNA is located 48 nt upstream of the 3' end of the
human genomic fragments which hybridized to an Endo B full-length cDNA. The placement of all K18 gene restriction
cDNA probe, the mouse equivalent of K18, under conditions sites found in the cDNA was consistent with a coding gene
that did not detect epidermal keratin sequences (71). Figure composed of multiple exons and introns. The coding region
1A shows the results of hybridizing the BstNI 5' riboprobe, of the gene is approximately 4.2 kb. Associated with it is 2.4
derived from the first 106 nt of the K18 cDNA, to restriction kb of upstream sequences and 3.3 kb of downstream se-
enzyme-digested HeLa cell DNA after washing the filter at quences.
55°C. The multiple bands may represent 15 to 20 related K18 To confirm the identity of the isolated gene, we sequenced
gene sequences. This result was very similar to that seen the 5' end of the coding region. The sequence of the 5' end
with an Endo B cDNA probe (71). Figure 1B shows the of the K18 gene from the XhoI site (nt -251) to the BamHI
pattern of the same filter after additional washing at 70°C. site (nt 1379) is shown in Fig. 3. The sequence containedVOL. 8, 1988 CLONING AND EXPRESSION OF HUMAN KERATIN 18 GENE 1543
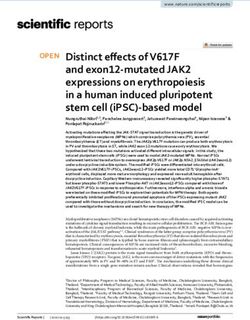
x Xm
H -252128 BllI B
;; ; NEO SV40 4
K18pO Neo I I
1kb
IH-H
BIl
1207
+1 mn B
H X 128 1379 HIl B B H
-2500 -252 464 2400 3100 4050 7450
;4l q e lJ1
K1 8 Gene
1kb ,'I ,,'1 f
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
1
I
F- 0
0
J,
,' 1245 , 4000
0
,p K
.0
XiMi B II P H II B K
128 464 503 745 1068 1357
v
K1 8 ctNA M-
0.1 Kb
l I
FIG. 2. Restriction maps of the K18 gene, cDNA, and K18pONeo expression vector. The map of the K18 gene is compared with that of
the cDNA. The restriction endonuclease sites shown are: X, XhoI; Xm, XmaIII; BII, BglII; P, PstI; B, BamHI; HII, HincII; K, KpnI; H,
HindIll. The 5' region (from XhoI at nt -252 to BamHI at nt 1379) of the K18 gene was labeled with reference to Fig. 3 in which +1 and +464
represent the first and last nucleotides of exon 1. The remaining portions are the estimated restriction fragment sizes. The double lines of
K18pONeo represent plasmid sequences of pGEM-1, the starting vector. The dark lines represent exons 1 and 2 of the K18 gene and the
cDNA. The BglII site of the K18 gene at nt 1207 overlaps the splice acceptor site of intron 1 and is reconstructed in the cDNA. Sites found
in both the gene and cDNA are connected by dotted lines. The top portion shows the structure of K18pONeo in which the neo gene and SV40
splice and polyadenylation signals from pSV2Neo were inserted at the BglII site of the cloned 5' BamHI fragment of the K18 gene.
exons 1 and 2, intron 1, and part of intron 2 as determined by present: one at bp - 140 to - 134, one at bp - 125 to - 119,
alignment of the sequence with the K18 cDNA sequence. and two tandem repeats between bp -65 and -51. No
Exon 1 is 464 bp (1 to 464); intron 1 is 741 bp (465 to 1205); typical CCAAT sequences (12) were found within this re-
exon 2 is 86 bp (1206 to 1293); and the partial intron 2 is 88 gion, but two were found within intron 1. The first is at bp
bp (1294 to 1381). The sequences of exons 1 and 2 are 509, and the second is inverted at bp 546. However, these
identical with the corresponding portion of the K18 cDNA are far from the normal positions found for such functional
except for the first four nucleotides of the published cDNA genes. The sequence CCACC, which may function as a
sequence. These four nucleotides appear to have been CCAAT box in humans, is found at nt -97. Also present in
derived from linkers originally added to the cDNA. The intron 1 is a possible binding site (TGGGAGGAGCCA) for
translational initiation (ATG) codon that begins the exon 1 nuclear factor I (NF-I) (27, 28) at bp 847 to 860.
open reading frame of the K18 gene is at nt 48 (Fig. 3). Exon Expression of K18 RNA in mouse cells. The recombinant
1 codes for the first 138 amino acids of the K18 protein, plasmid pGC1853, which contains the entire K18 gene (4.2
which includes the head region, coil 1A, spacer 1, and the kb) along with approximately 2.4 kb of 5' upstream sequence
first 11 amino acids of the coil 1B protein domains. Exon 2 and approximately 3.3 kb of 3' downstream sequence, was
codes for the next 29 amino acids of coil 1B. The size of exon cotransfected with pSV2Neo into F9.22 embryonal carci-
1 (464 bp) is slightly smaller than the 525-bp average exon 1 noma cells, HR9 murine parietal endodermal cells, and STO
of type I keratins, while exon 2 (86 bp) is close to the average and L-cell mouse fibroblasts. One of five F9 clones, four of
size (84 bp) for type I keratins (41). Intron 1 (740 bp) is four HR9 clones, two of five STO clones, and four of four
considerably smaller than the 1,256-bp size of epidermal L-cell clones expressed an immunoprecipitable amount of a
type I keratins. The sequence of the donor splice junction for K18-like protein. F9 clones which express K18 are appar-
exon 1 is AGGTAAGG, while that for exon 2 is AGT ently quite rare because further analysis of a pooled popu-
CAAGT. Introns 1 and 2 each begin with a GT, and intron 1 lation of approximately 100 additional clones showed no
ends with an AG. These are highly conserved sequences at detectable K18-like protein (data not shown). Considering
the splice junction for most eucaryotic mRNAs (51). The the sensitivity of the assay, we estimate that fewer than 1 in
transcriptional start site was mapped by the Si technique every 20 transfected F9 clones expresses K18.
(see below) and was within 4 bp of the start of the previously Si analysis of total cellular RNA was performed to deter-
isolated cDNA. mine the start site of the K18 mRNA in these stably
The 252-bp region upstream of the transcriptional start site transfected cell lines. The probe for K18 mRNA was ex-
has a high G+C content (71%) with a TATA-like box pected to protect a fragment of approximately 240 nt.
embedded within the 5' region at bp -38 to -43. Four Similarly, approximately 195 nt of the Endo B probe were
possible SPl-binding sites (GGGGCGG) (22) are also expected to overlap with the 5' end of Endo B mRNA. The1544 KULESH AND OSHIMA MOL. CELL. BIOL.
-252 5' cctcgagc casaWKCc tgctgtccgt gtccatgccc ggttggecc cccgtttctg
-192 ggggtgwgc ggggcttggc sggtgwgcghaghggcutggc ccgc gcggagggcg cgggctccp gccgtccacc
................. .........
-92 tgtggctceg gcttccga egctccg gcgg cg guectcectc tgcgstates ctcggtcgc gcgctcgcg coggccgcca cstcstcs
9 casectas atectatect ttctctctcc e e tsmuttcc c ctcsatcc acttctcca cseetacca atecetaocc tctatcca
109 cacceact ec cct e c.atctataca maCt s ctcttttc ccatctcc atatcccact ccaccaatt
209 ecaa at_otcc_ acctac
ecte tc t _ atc caacs .aamacat saaccsta
309 -e------ -a ctta cctan at_mmce tac amcca ctaaaaa ttaaa aaaaaa.c
409 aatcatca aaactat actamtas ggtaa gacctcaa tcccawctt gtctgacct
r_-=3) (33333
509 ccaattatac actcctttgc ctctttccgt cattccata ccaceccasc ccctactcca ccgggag gttg.cata cctuatttc catccgcgca
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
609 cctaWcaca ggtcccta ccc tagcat ghhagw tc tttccca g _gg a cgg gttgagat ttacagagp
709 *gtggcWc atagggag gtaagaa gwettaws actggctctg gcggtgn ctattgg mttaagcs gatgtggcta
000000000
809 aggctgtc atetaggagt aaacagag ccttcctttg _gagga atcca tg taghucc agtacca gtgcactag ggaaatg
909 ccag_g"a goccaggg ggcttgtta gtagcgmtc acttctgc aggcagcc gcagctaw cagcctgctg aggttccca agagcag
1009 agtgctng tctggatc caggaagg gggatgggg tgggtag tgaag taggtgtcca gggea ct ctggctattc ctgggaccag
1109 gaagttttca ctaptac taacatttt tacactca ccccceeat ccctggcttt ctattcatg aaccctct ctctacatc cctccagg
1209 ttcscaata CtatascWa. tc cat
O ttCtact=a ttacaatc Occtttact actaatact ttaatca ataatttg ggct"ag
1309 gctggggtc cagtap gctaaga actgctcccc agctggta gttaggce c"atg tcc 3'
FIG. 3. Nucleotide sequence of exons 1 and 2 and the 5'-flanking region of the human K18 gene. The DNA sequence is listed 5' to 3', left
to right, and is the same sense as the K18 mRNA sequence. The entire sequence was independently determined on both strands of the cloned
3.9-kb 5' fragment. The transcriptional start site of the K18 mRNA, determined by S1 nuclease protection analysis, is indicated by carets (^^).
Exon 1 (bp 1 to 464) and exon 2 (bp 1206 to 1293) are underlined and are identical with the corresponding portion of the K18 cDNA. The
translation initiation codon is marked by the plus signs (+ + +) above the ATG. Intron 1 (741 bp) and the partial intron 2 (88 bp) are the
nonunderlined sequences after exons 1 and 2. Within intron 1, double broken lines (= = = =) mark two CCAAT sequences, while open circles
(0000) designate a possible NF-I-binding site. The 5'-flanking region contains an ATATAA sequence marked by the asterisks (****) at bp
-38 to -43 and four possible SPi-binding sites (GGGGCCGG) designated by overlines (-) at bp -140, -125, and a tandem repeat between
bp -65 and -51.
first 250 bases of the K18 cDNA are only 59% identical with 9), one other major fragment of 190 bases appears to map
the Endo B cDNA (56) with no identical stretch of greater very close to the ATG (nt 48) translation start site and some
than 32 nt. Therefore, the probes should only protect their minor start sites immediately upstream of the expected one
respective mRNAs. Figure 4A demonstrates both the spec- were found. The significance of these secondary protected
ificity and sensitivity of the K18 probe in the Si nuclease fragments is currently under further investigation.
protection assay. The size of the Si-protected fragment Duplicate samples of total cellular RNA were also hybrid-
derived from hybridization with HeLa cell or BeWo tropho- ized to the Endo B probe, which protected an Endo B
blastoma RNA and digestion with Si nuclease was 240 mRNA fragment of 195 nt from HR9 cells (Fig. 4B, lane 12).
bases, as expected (lanes 1 to 6). (See Fig. 3 for the location This fragment maps the major mRNA start site to within 5 nt
of this site within the K18 gene sequence.) K18 mRNA was of the start of the cDNA sequence as previously suggested
detectable in as little as 0.05 ,ug of total RNA from HeLa by primer extension analysis (71). No Endo B signal was
cells (lane 1). The same size fragment was found in the L-3 detected in RNA from HeLa cells (lane 6), L-3 cells (lane 8),
cells which had been transfected with the K18 gene (lane 8) F9-3 cells (lane 10), or STO-6 cells (lane 16). Therefore,
but not in control L cells (lane 7). The K18 mRNA produced correct transcription of the K18 gene can occur in cell types
in L-3 cells has the same transcriptional start site as that that do not express the homologous Endo B gene. The
found in HeLa and BeWo cells, and its abundance was transfected HR9 cells express both K18 and Endo B simul-
approximately 25% of that of HeLa cells. Yet, this cell line taneously (compare lane 13 with lane 14).
accumulated less than 5% of the K18 protein found in HeLa To determine whether the SV40 promoter or enhancer
cells (see Fig. 5A, lane 10). elements present in the cotransfected pSV2Neo plasmid had
Figure 4B shows the results of mapping the 5' ends of both an influence on the expression of the K18 gene, the neo gene
K18 and Endo B RNAs in additional cell lines. Lanes 5 to 20 and the downstream SV40-derived splice and polyadenyla-
represent the Si analysis of total RNA from the various tion signals from pSV2Neo were inserted into the BglII site
transfected cell lines. The samples shown in the odd-num- of the pGC39 plasmid which contains only the 3.9-kb
bered lanes were hybridized to the K18 probe, and those HindIII-to-BamHI fragment of the 5' end of the K18
shown in the even-numbered lanes were hybridized to the gene. Figure 2 shows the structure of this construction
Endo B probe. HeLa RNA (lanes 5 and 6) and HR9 RNA (K18pONeo). The resulting plasmid was transfected into
(lanes 11 and 12) were used to identify the representative HR9 cells, L cells, and mouse EC cell lines. Colonies
Si-protected fragments(s) for K18 and Endo B RNA, respec- resistant to the antibiotic G418 were found at a frequency of
tively. The K18 protein-synthesizing cells all show a major approximately 2.3 x 10-4, 3 x 10-5, and 3 x 1i-5 in L cells,
Si-protected fragment of 240 nt (lanes 5, 7, 9, 13, and 15), HR9 cells, and HeLa cells, respectively, but at only 1 x 10-6
indicating a proper initiation of transcription within the in F9 EC cells. These results are consistent with the very low
transfected K18 gene. In the sample from F9-3 (Fig. 4B, lane frequency with which F9 cells express the entire transfectedVOL. 8, 1988 CLONING AND EXPRESSION OF HUMAN KERATIN 18 GENE 1545
0
0q
A. B.
M l 2 3 4 56 7 8 M 1 2 '3 4'5 6'7 8 9 1011 121314i51617181192O
818- -
652- -
10
472545 -
454_-.-- ii.
357- m4
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
_
we_ 4240 No
4240
193- *.
176-
156-
_l ~~t al *_ 4195
129-
123- -
111- -
FIG. 4. S1 analysis of K18 and Endo B RNA. The 5' ends of the K18 mRNA and Endo B mRNA were located by S1 nuclease protection
of the 32P-labeled K18 Si probe and the Endo B Si probe, respectively, as described in Materials and Methods. (A) Lane M, Marker
32P-end-labeled fragments of HpaII-digested M13mpl9-RF. The nucleotide sizes of the fragments are indicated on the left. Lane 1, 0.05 ,ug;
lane 2, 0.1 ,ug; lane 3, 0.5 ,ug; lane 4, 1.0 ,g; lane 5, 5.0 j,g of total RNA from HeLa cells. Lane 6, 1.0 pg of total RNA from BeWo cells (56).
Lane 7, 5.0 ,ug of total RNA from mouse L cells. Lane 8, 5.0 ,ug of total RNA from L-3 cells. All samples were hybridized to 10' cpm of the
K18 S1 probe, digested with S1 nuclease, resolved on a sequencing gel, and detected by autoradiography in the presence of an enhancing
screen for 48 h. (B) Lane M, Marker fragments as for panel A. Lane 1, Undigested K18 Si probe (471 nt). Lane 2, Undigested Endo B S1
probe (780 nt). Lanes 3 and 4, 5.0 ,ug of tRNA. Samples of odd-numbered lanes (5 to 19) received 5.0 ,ug of total RNA isolated from the
indicated cell line hybridized to 5 x 104 cpm of the K18 S1 probe. Even-numbered lanes (6 to 20) are duplicate RNAs hybridized to 5 x 104
cpm of the Endo B S1 probe. Lane 21, Sanger sequencing reaction for cytosine of the Endo B probe used for additional size standards.
Numbers on the right side indicate the sizes of the major S1-protected fragments. All samples were analyzed by electrophoresis on an 8 M
urea-6% acrylamide gel followed by autoradiography. Panel B was exposed for 96 h, except for lanes 1, 2, and 9, which were exposed for
48 h.
K18 gene and suggest that F9 cells do not utilize the K18 the K18 protein. Duplicate cell lysates were immunoprecip-
promoter efficiently. Sl protection analysis of the repre- itated with Endo A antiserum, which also recognizes K8
sentative G418-resistant clones of L cells and HR9 cells is (Fig. 5B). Note that lanes 1 to 8 were exposed for 3 days,
shown in Fig. 4B, lanes 17 to 20. The K18 probe is fully while lanes 9 to 11 were exposed for 14 days owing to the low
contained within the K18 portion of the neo construction and amount of signal present in the L cells. HeLa cells, which
thus would be expected to protect the same size fragment as express endogenous K18 (Fig. SA, lane 1) and K8 (Fig. 5B,
authentic K18 RNA. Inspection of lanes 17 and 19 indicates lane 1), and mouse HR9 cells, which express Endo B (Fig.
that the 5' end of the K18 gene (5'-flanking region, exon 1, 5A, lane 5) and Endo A (Fig. SB, lane 5), were positive
and intron 1) is sufficient to generate an appropriately controls. The human forms of these keratins could be
initiated mRNA in the absence of SV40 promoter and distinguished from their mouse counterparts by their elec-
enhancer sequences. trophoretic mobility (Fig. 5A and B, lanes 1 and 5). The
Expression of K18 and Endo B proteins in mouse cells. mouse STO, F9 EC, and L cells do not synthesize either
Immunoprecipitation and subsequent partial peptide map- Endo A or B. In the transfected cells which had been
ping were performed both to verify the identity of the K18 exposed to [35S]methionine for a relatively long period (Fig.
protein and to analyze the effect of its expression on the 5A), STO-6 (lane 4) and HR9-3 (lane 6) accumulated approx-
endogenous Endo A and Endo B proteins. Figures SA and B imately 30% of the K18 found in HeLa cells (Fig. SA, lane 1),
show the results of immunoprecipitation of several transfec- while HR9-5 (lane 7) and HR9-8 (lane 8) accumulated ap-
ted cell lines. The cell lysates in Fig. 5A were immunopre- proximately 40 and 60%, respectively, of the K18 found in
cipitated with Endo B antiserum, which cross-reacts with HeLa cells. Both L-3 and L-8 (lanes 10 and 11, respectively)1546 KULESH AND OSHIMA MOL. CELL. BIOL.
A. C.
Endo B -_
Keratin 18 68,000-
43,000-
1 2 3 4 5 6 7 8 9 1011
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
.4wo:
B. -or-0 .ICbelb Kb'Of.b VW
,.Pb. 84Nq '4.. \e NV
CO .o ", Oa -NNW
-
Endo A-.
"
Keratin 8'
.7
1 2 3 4 5 6 7 8 9 1011
7,600> 40 oft
1,500-
1 2 3 4
FIG. 5. Immunoprecipitation and peptide mapping of the K18 protein from stably transfected cell lines. The indicated cell lines were
cotransfected with pGC1853 and pSV2Neo. Stably transfected cell lines were selected in medium containing G418 (400 ,ug/ml).
[35S]methionine-labeled (15 h) whole-cell lysates (107 cpm) of both control cells (lane 1, HeLa; lane 2, STO mouse fibroblasts; lane 5, HR9
mouse parietal endodermal cells; lane 9, L cells) and their stably transfected counterparts (lanes 3 and 4; 6, 7, and 8; 10 and 11) were
immunoprecipitated with either Endo B (A) or Endo A (B) antiserum and analyzed by SDS-acrylamide gel electrophoresis and fluorography.
Lanes 1 to 8 were exposed for 3 days, while lanes 9 to 11 were exposed for 14 days. The positions of Endo B, K18, Endo A, and K8 are
indicated on the left. (C) Partial peptide mapping of immunoprecipitated Endo B from HR9 cells and K18 from either HeLa cells or L cells
(clone L-3) and F9 cells (clone F9-3) that had received pGC1853. Immunoprecipitates were digested with trypsin (final concentration of 0.6
,ug/ml) at 37°C for 15 min before being loaded on an SDS-acrylamide gel. Protein fragments were detected by fluorography. Lane 1, Control
Endo B protein from HR9 cells; lane 2, control K18 protein from HeLa cells; lanes 3 and 4, immunoprecipitated protein from stably
transfected F9 EC cells and L cells, respectively. Molecular weight markers are indicated on the left. The undigested immunoprecipitate of
F9-3 cells (data not shown) was very similar to that of L-3 cells shown in panel A (also see Fig. 6, lane 6).
accumulated less than 5% of the total HeLa K18 protein, lanes 6 to 8) resulted in the accumulation of compensatingly
while F9-3 (data not shown) contained about 15%. It is decreasing amounts of Endo B. Table 1 summarizes the
evident that fibroblasts (L cells and STO cells) which do not relative levels of K18, Endo B, and Endo A in these cells. In
express the homologous Endo B gene can express the HR9-5 and HR9-8 cells, the sum of K18 and Endo B protein
transfected K18 gene. was nearly equal to that found in control HR9 cells. The
The K18 protein expressed by the transfected cell lines level of Endo A was the same in all three lines. In HR9-3
was compared with that made by HeLa cells and with Endo cells, the level of Endo A was higher, but the sum of K18 and
B by proteolytic digestion and gel electrophoresis. The Endo B was also slightly higher and was within the range
peptide pattern of the immunoprecipitated protein in F9-3 found for randomly selected subclones of HR9. Thus, it
(Fig. SC, lane 4) and L-3 (lane 3) cells was the same as that
of HeLa cell K18 (lane 2) and distinctly different from that of
Endo B (lane 1). The fourth fragment from the top, seen in TABLE 1. Relative levels of K18, Endo A, and Endo B
lane 3, is a degradation intermediate also found in shorter in stably transfected cell linesa
digests of the other two samples and not present with slightly Cell line K18 Endo B Endo A )18 + Endo
longer digestion times (data not shown). These results, in
addition to the size of the undigested protein and the HR9 0.0 1.0 1.0 1.0
sequence data, confirm that the correct K18 gene was HR9-3 0.3 1.0 1.2 1.1
isolated. HR9-5 0.3 0.7 1.0 1.0
The expression of K18 did not appreciably affect the levels HR9-8 0.5 0.6 1.0 1.1
of Endo A in HR9 cells (Fig. SB) nor did it induce the
expression of this normally silent type II keratin in STO or a Radioactivity of the samples shown in Fig. 4A and B, lanes 5, 6, 7, and 8,
was quantitated by densitometry with appropriately exposed films. The values
L-cell fibroblasts. However, in contrast to Endo A, expres- of K18 and Endo B are arbitrary units set to 1.0 for Endo B in the HR9 cells.
sion of increasing amounts of K18 in HR9 cells (Fig. SA, Endo A values are relative to Endo A in the HR9 cells.VOL. 8, 1988 CLONING AND EXPRESSION OF HUMAN KERATIN 18 GENE 1547
Hela L-3 F9-3 HR9 HR9-3 HR9-8 isolation and expression of the gene for K18 provides
information concerning both these questions.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Southern analysis under conditions which do not detect
epidermal keratin or other IF genes indicates that the human
genome contains at least 15 to 20 genes similar to K18 (71)
__ Endo B (Fig. 1). This is in contrast to the five Endo B genes and two
_
___ _ Ker 18 Endo A genes found in mouse DNA (65, 71, 72). The larger
number of K18 genes in humans may be due to the expres-
sion of K18 in very early embryonic stem cells which could
FIG. 6. Stability of the K18 protein. [35S]methionine-labeled lead to virus-mediated generation of pseudogenes (37). The
whole-cell lysates were prepared from 1.5-h-labeled cells (lane 1, expression of keratins in human EC cells (19) but not mouse
HeLa; lane 3, L-3; lane 6, F9-3; lane 9, HR9; lane 11, HR9-3; lane EC cells is consistent with this hypothesis. Epidermal kera-
14, HR9-8) and from duplicate plates labeled for 1.5 h and chased tins which are expressed much later in development are
with cold methionine in Dulbecco modified Eagle medium (see considered to be primarily single-copy genes (41). To date,
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
Materials and Methods) for either 3 h (lane 4, L-3; lane 7, F9-3; lane we have found no data indicating more than one active Endo
12, HR9-3; lane 15, HR9-8) or 9 h (lane 2, HeLa; lane 5, L-3; lane 8, B or K18 gene (56, 71). Several lines of evidence indicate
F9-3; lane 10, HR9; lane 13, HR9-3; lane 16, HR9-8). Each lysate (5 that we succeeded in isolating the active K18 gene from the
x 106 cpm) was immunoprecipitated with Endo B antiserum and many related sequences in the human genome. The se-
subjected to electrophoresis and fluorography for 5 days. quences of exons 1 and 2 correspond exactly to that of the
previously determined cDNA including the 5' noncoding
appears that the steady-state level of endogenous Endo B leader (Fig. 3). Transfection of the K18 gene resulted in the
can be modulated to accommodate the expression of rela- expression of a protein which was indistinguishable from
tively high levels of K18. authentic K18 in antigenicity, electrophoretic mobility, and
The relatively abundant K18 mRNA found in L-3, F9-3, partial peptide pattern (Fig. 4). The RNA derived from either
and STO-6 cells (Fig. 4A, lane 8, and Fig. 4B, lanes 7, 9, and the transfected gene or a recombinant construction which
15) compared with the relatively low levels of K18 protein drives the bacterial neo gene was initiated at the same
found in the same cells after labeling overnight (Fig. 5A, position as authentic K18 mRNA. Thus, the cloned 9.9-kb
lanes 4 and 10) suggested that the K18 protein was turning genomic fragment contains all necessary regulatory se-
over more rapidly in cells which do not contain Endo A or quences required for faithful transcriptional initiation, splic-
another type II keratin with which it could form intermediate ing, and accurate translation.
filaments. The stability of the K18 protein in both control The sequence of the K18 gene upstream of the transcrip-
and transfected cells was determined by labeling the cells for tional initiation site is very different from that of other IF
1.5 h with [35S]methionine medium followed by a chase in genes coding for vimentin (60), desmin (61) or epidermal
complete nonradioactive medium for 3 or 9 h and analysis by keratins (10, 42). However, several small sequence motifs
immunoprecipitation. After a relatively short labeling time, important for the regulation of other eucaryotic promoters
the amount of radioactive K18 found in either L-3 or F9-3 were found. A TATA box (12) is found at nt -38. Four
cells was much more comparable to that in HeLa cells than potential SP1 (22, 40)-binding sites (GGGGCGG) were
when an overnight labeling was performed (compare Fig. 6, found, and intron 1 also contained a potential NF-I-binding
lanes 1, 3, and 6, with Fig. 5A, lanes 1 and 10, and the total site at nt 847 to 860. NF-I is required for the efficient in vitro
recovered signal in lane 4 of Fig. SC). In contrast, the replication of adenovirus (50) and appears to be involved in
differences in the levels of Endo B in HR9-8 and HR9 control the selective recognition of some eucaryotic promoters (33).
cells labeled overnight (Fig. SA, lanes 5 and 8; Table 1) were NF-I-binding sites found in HeLa cells are considered to be
confirmed even when the much shorter labeling period was present within single-copy DNA (approximately 1 per 100
used (Fig. 6, lanes 9 and 16). This suggests that Endo B kb) (27, 28). It is of interest that NF-I-binding activity was
protein synthesis is down regulated by the expression of not detected in F9 cells (67). Determination of the functional
K18. After chasing for either 3 or 9 h, K18 in L-3 and F9-3 significance of these K18 sequences will require additional
cells was greatly diminished (Fig. 6, lanes 4, 5, and 7, 8). investigation.
However, K18 was stable in HeLa cells or in transfected The 5'-flanking region of the K18 gene is G+C rich (71%),
HR9 cells (Fig. 6, lanes 1, 2, 12, 13, 15, and 16). It appears and the dinucleotides CG and GC are roughly equal in
that K18 is stable only in cells which express a compatible number (35 and 39, respectively). Thus, this region could be
type II keratin (either K8 or Endo B) with which it can form considered an HpaII tiny fragment (HTF)-like island. HTF
intermediate filaments. islands are recently described G+C-rich regions of under-
methylated genomic DNA which contain relatively high
DISCUSSION levels of the normally underrepresented dinucleotide CG (9).
Most such regions are unique in sequence and are associated
The mouse form of K18, Endo B, along with its type II with genes which are constitutively active. The methylation
keratin partner, Endo A, appear to be the first IF proteins to status of the HTF-like island of K18 has not yet been
be expressed during embryonic development (13, 31, 55). In determined. However, the possibility that methylation of
the adult, the expression of K18 is limited primarily to simple this region is involved in the regulation of this gene, which is
epithelial cell types of diverse specialized functions. What expressed in many but not all differentiated cells, is intrigu-
regulatory mechanisms are involved in limiting the expres- ing. Consistent with this notion is the observation that the
sion of K18 in many but certainly not all differentiated cells? expression of Endo B, the mouse form of K18, can be
Moreover, because keratin IF formation requires two dif- induced in myoblasts, which do not normally express the
ferent subunits, how are the levels of the two different gene, by treatment with azacytidine, an agent which results
subunits balanced to produce the apparently equal molar in reduced DNA methylation (20).
amounts of type I and II keratins found in filaments? The Transfection of either the K18 gene of the K18pONeo1548 KULESH AND OSHIMA MOL. CELL. BIOL.
construction results in relatively efficient expression in both This investigation was supported by Public Health Service grants
HR9 endodermal cells and fibroblasts as judged by either the CA 42303 and CA 33946 from the National Cancer Institute and by
colony numbers or steady-state levels of RNA in selected training grant T32 CA 09497 from the National Institutes of Health.
clones (Fig. 4). It also suggests that fibroblasts do not
contain high concentrations of a trans-acting negative regu- LITERATURE CITED
latory activity that can suppress K18 expression. The 1. Andrews, P. W., D. L. Bronson, F. Benham, S. Strickland, and
expression of K18 in cells which do not normally express B. B. Knowles. 1980. A comparative study of eight cell lines
keratins did not result in the activation of the endogenous derived from human testicular teratocarcinoma. Int. J. Cancer
26:269-280.
Endo A or Endo B genes. This result is similar to that 2. Andrews, P. W., P. N. Goodfellow, and I. Damjanov. 1983.
reported for the desmin gene, which can be expressed in Human teratocarcinoma cells in culture. Cancer Surv. 2:41-73.
cells which do not express their endogenous desmin gene 3. Bader, B. L., T. M. Magin, M. Hatzfeld, and W. W. Franke.
(61). In addition, it is consistent with the recent report that 1986. Amino acid sequence and gene organization of cytokeratin
the forced expression of a human type I epidermal keratin no. 19, an exceptional tail-less intermediate filament protein.
cDNA in mouse cells does not induce endogenous type II EMBO J. 5:1865-1875.
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
expression, although the converse may occur (26). 4. Barklis, E., R. C. Mulligan, and R. Jaenisch. 1986. Chromo-
In contrast to the results of transfecting endodermal cells somal position or virus mutation permits retrovirus expression
or fibroblasts, very few F9 cells expressed either the K18 in embryonal carcinoma cells. Cell 47:391-399.
5. Barr, P. J., R. M. Thayer, P. Laybourn, R. C. Najarian, F.
gene or the K18pONeo vector. This result was not due to Seela, and D. R. Tolan. 1986. 7-deaza-2'-deoxyguanosine-5'-
differences in the efficiency of transfection, because in the triphosphate: enhanced resolution in M13 sequencing. BioTech-
case of cotransfection of the K18 gene with pSV2Neo, niques 4:428-432.
hundreds of G418-resistant clones were obtained but only 6. Benton, W. D., and R. W. Davis. 1977. Screening lambda gt
one expressed the K18 mRNA and protein. This clone, F9-3, recombinant clones by hybridization to single plaques in situ.
was the only one which showed relatively large amounts of Science 196:180-182.
additional and unusual 5' ends of the K18 mRNA (Fig. 4, 7. Bernstine, E. G., M. L. Hooper, S. Grandchamp, and B.
lane 9). In addition, it was unusual in that it expressed the Ephrussi. 1973. Alkaline phosphatase activity in mouse tera-
highest level of K18 RNA of all the transfected clones toma. Proc. Natl. Acad. Sci. USA 70:3899-3903.
8. Biggin, M. D., T. J. Gibson, and G. F. Hong. 1983. Buffer
analyzed. We suggest that the K18 gene does not work gradient gels and 35S label as an aid to rapid sequence determi-
efficiently in F9 cells, although rare and somewhat unusual nation. Proc. Natl. Acad. Sci. USA 80:3963-3965.
clones can be isolated. This rare expression of the transfec- 9. Bird, A. 1986. CpG-rich islands and the function of DNA
ted K18 gene in F9 EC cells may be due to particular sites of methylation. Nature (London) 321:209-213.
integration as has been documented for the rare utilization of 10. Blessing, M., H. Zentgraf, and J. L. Jorcano. 1987. Differentially
retroviral promoters in EC cells (4). Thus, F9 cells which do expressed bovine cytokeratin genes. Analysis of gene linkage
not express the homologous Endo B gene but can be induced and evolutionary conservation of 5'-upstream sequences.
to express it upon differentiation to extraembryonic endo- EMBO J. 6:567-575.
derm (53) differ from fibroblasts which also do not express 11. Bonner, W. M., and R. A. Laskey. 1974. Film detection method
for tritium-labelled proteins and nucleic acids in polyacrylamide
Endo B. F9 cells also differ from fibroblastic type cells in gels. Eur. J. Biochem. 46:83-88.
that azacytidine treatment does not result in the induction of 12. Breathnach, R., and P. Chambon. 1981. Organization and
Endo B in F9 cells (unpublished data). expression of eukaryotic split genes coding for proteins. Annu.
At the protein level, the modest levels of K18 which Rev. Biochem. 50:349-383.
accumulate in transfected F9 cells and L-cell fibroblasts 13. Brulet, P., C. Babinet, R. Kemler, and F. Jacob. 1980. Mono-
appear to be due to relatively rapid turnover of the protein clonal antibodies against trophectoderm-specific markers during
(Fig. 6). This is in contrast to the stability of K18 in mouse blastocyst formation. Proc. Natl. Acad. Sci. USA
transfected HR9 cells, of the endogenous Endo B in HR9 77:4113-4117.
cells, and of K18 in human mesothelial cells (17). The 14. Brulet, P., P. Duprey, M. Vasseur, M. Kaghad, D. MoreOlo, P.
Blanchet, C. Babinet, and H. Condamine. 1985. Molecular
stability of K18 in HR9 cells suggests that the expression of analysis of the first differentiations in the mouse embryo. Cold
a type II keratin with which K18 could polymerize is a Spring Harbor Symp. Quant. Biol. 1:51-57.
sufficient condition for stabilizing K18. A direct test of this 15. Chung, A. E., L. E. Estes, H. Shinozuka, J. Braginski, C. Lorz,
hypothesis will require the isolation and expression of an and C. A. Chung. 1977. Morphological and biochemical obser-
appropriate type II keratin in fibroblasts already expressing vations on cells derived from the in vitro differentiation of the
unstable K18. embryonal carcinoma cell line PCC4-F. Cancer Res.
While the expression of K18 in HR9 cells appears to have 37:2072-2081.
little or no effect on the expression of endogenous Endo A 16. Cleveland, D. W., S. G. Fischer, M. W. Kirschner, and U. K.
(Fig. SB; Table 1), it does result in the modulation of the Laemmli. 1977. Peptide mapping by limited proteolysis in so-
dium dodecyl sulfate and analysis by gel electrophoresis. J.
level of Endo B (Fig. SA; Fig. 6, lanes 11 and 14; Table 1). Biol. Chem. 252:1102-1106.
Competition of K18 with Endo B for Endo A subunits and 17. Connell, N. D., and J. G. Rheinwald. 1983. Regulation of the
subsequent turnover of excess K18 or Endo B may contrib- cytoskeleton in mesothelial cells: reversible loss of keratin and
ute to the lower level of Endo B protein. However, the lower increase in vimentin during rapid growth in culture. Cell
level of Endo B is also due, at least in part, to a lower rate of 34:245-253.
Endo B protein synthesis (Fig. 5A). Determination of 18. Damjanov, I., and P. W. Andrews. 1983. Ultrastructural differ-
whether this effect is regulated transcriptionally or posttran- entiation of a clonal human embryonal carcinoma cell line in
scriptionally will be of particular interest. vitro. Cancer Res. 43:2190-2198.
19. Damjanov, I., R. K. Clark, and P. W. Andrews. 1985. Expres-
ACKNOWLEDGMENTS sion of keratin polypeptides in human embryonal carcinoma
cells, p. 732-733. In E. Wang, D. Fischman, R. K. H. Liem, and
We thank Grace Cecena for her excellent technical expertise and T. T. Sun (ed.), Intermediate filaments. New York Academy of
Tina Trevor, Beth Mather, Frank Fujimura, Terumi Kohwi-Shige- Sciences, New York.
matsu, and Eileen Adamson for their comments on the manuscript. 20. Darmon, M., J. F. Nicolas, and D. Lamblin. 1984. 5-AzacytidineVOL. 8, 1988 CLONING AND EXPRESSION OF HUMAN KERATIN 18 GENE 1549
is able to induce the conversion of teratocarcinoma-derived sequence of a gene encoding a human type I keratin: sequences
mesenchymal cells into epithelial cells. EMBO J. 3:961-967. homologous to enhancer elements in the regulatory region of the
21. Davis, L. G., M. D. Dibner, and J. F. Battey. 1986. Basic gene. Proc. Natl. Acad. Sci. USA 82:1609-1613.
methods in molecular biology, p. 276-284. Elsevier Science 43. Martin, G. R., and M. J. Evans. 1975. Differentiation of clonal
Publishing Co., New York. lines of teratocarcinoma cells: formation of embryoid bodies in
22. Dynan, W. S., and R. Tjian. 1985. Control of eukaryotic vitro. Proc. Natl. Acad. Sci. USA 72:1441-1445.
messenger RNA synthesis by sequence-specific DNA-binding 44. McKeon, F. D., M. W. Kirschner, and D. Caput. 1986. Homol-
proteins. Nature (London) 316:774-778. ogies in both primary and secondary structure between nuclear
23. Eichner, R., P. Bonitz, and T. Sun. 1984. Classification of envelope and intermediate filament proteins. Nature (London)
epidermal keratins according to their immunoreactivity, isoelec- 319:463-468.
tric point and mode of expression. J. Cell Biol. 98:1388-1396. 45. Meinkoth, J., and G. Wahl. 1984. Hybridization of nucleic acids
24. Franke, W. W. 1987. Nuclear lamins and cytoplasmic interme- immobilized on solid supports. Anal. Biochem. 138:267-284.
diate filament proteins: a growing multigene family. Cell 48:3-4. 46. Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K.
25. Franke, W. W., E. Schmid, S. Winter, M. Osborn, and K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of
Weber. 1979. Widespread occurrence of intermediate-sized fil- biologically active RNA and RNA hybridization probes from
aments of the vimentin-type in cultured cells from diverse plasmids containing a bacteriophage SP6 promoter. Nucleic
Downloaded from http://mcb.asm.org/ on January 14, 2021 by guest
vertebrates. Exp. Cell Res. 123:25-46. Acids Res. 12:7035-7055.
26. Giudice, G. J., and E. Fuchs. 1987. The transfection of epider- 47. Messing, J. 1983. New M13 vectors for cloning. Methods
mal keratin genes into fibroblasts and simple epithelial cells: Enzymol. 101:20-78.
evidence for inducing a type I keratin by a type II gene. Cell 48. Mizusawa, S., S. Nishimura, and F. Seela. 1986. Improvement of
48:453-463. the dideoxy chain termination method of DNA sequencing by
27. Gronostajski, R. M., S. Adhya, K. Nagata, R. A. Guggenheimer, use of deoxy-7-deazaguanosine triphosphate in place of dGTP.
and J. Hurwitz. 1985. Site-specific DNA binding of nuclear Nucleic Acids Res. 14:1319-1324.
factor I: analysis of cellular binding sites. Mol. Cell. Biol. 49. Moll, R., W. W. Franke, D. L. Schiller, B. Geiger, and R.
5:964-971. Krepler. 1982. The catalog of human cytokeratins: patterns of
28. Gronostajski, R. M., K. Nagata, and J. Hurwitz. 1984. Isolation expression in normal epithelia, tumors and cultured cells. Cell
of human DNA sequences that bind to nuclear factor I, a host 31:11-24.
protein involved in adenovirus DNA replication. Proc. Natl. 50. Nagata, K., R. A. Guggenheimer, T. Enomoto, J. H. Lichy, and
Acad. Sci. USA 81:4013-4017. J. Hurwitz. 1982. Adenovirus DNA replication in vitro: identi-
29. Hatzfeld, M., and W. W. Franke. 1985. Pair formation and fication of a host factor that stimulates synthesis of the preter-
promiscuity of cytokeratins: formation in vitro of heterotypic minal protein-dCMP complex. Proc. Natl. Acad. Sci. USA
complexes and intermediate-sized filaments by homologous and 79:6438-6442.
heterologous recombinations of purified polypeptides. J. Cell 51. Ohshima, Y., and Y. Gotoh. 1987. Signals for the selection of a
Biol. 101:1826-1841. splice site in pre-mRNA: computer analysis of splice junction
30. Henikoff, S. 1984. Unidirectional digestion with exonuclease III sequences and like sequences. J. Mol. Biol. 195:247-259.
creates targeted breakpoints for DNA sequencing. Gene 52. Oshima, R. G. 1978. Stimulation of the clonal growth and
28:351-359. differentiation of feeder layer dependent mouse embryonal
31. Jackson, B. W., C. Grund, E. Schmid, K. Burke, W. Franke, carcinoma cells by beta-mercaptoethanol. Differentiation
and K. Illmensee. 1980. Formation of cytoskeletal elements 11:149-155.
during mouse embryogenesis. Intermediate filaments of the 53. Oshima, R. G. 1981. Identification and immunoprecipitation of
cytokeratin type and desmosomes in preimplantation embryos. cytoskeletal proteins from murine extra-embryonic endodermal
Differentiation 17:161-179. cells. J. Biol. Chem. 256:8124-8133.
32. Johnson, L. D., W. W. Idler, X. Zhou, D. R. Roop, and P. M. 54. Oshima, R. G. 1982. Developmental expression of murine
Steinert. 1985. Structure of a gene for the human epidermal extraembryonic endodermal cytoskeletal proteins. J. Biol.
67-kDa keratin. Proc. Natl. Acad. Sci. USA 82:1896-1900. Chem. 257:3414-3421.
33. Jones, K. A., J. T. Kadonaga, P. J. Rosenfeld, T. J. Kelly, and R. 55. Oshima, R. G., W. E. Howe, F. G. Klier, E. D. Adamson, and
Tjian. 1987. A cellular DNA-binding protein that activates L. H. Shevinsky. 1983. Intermediate filament protein synthesis
eukaryotic transcription and DNA replication. Cell 48:79-89. in preimplantation murine embryos. Dev. Biol. 99:447-455.
34. Krieg, T. M., M. P. Schafer, C. K. Cheng, D. Filpula, P. 56. Oshima, R. G., J. L. Millan, and G. Cecena. 1986. Comparison
Flaherty, P. M. Steinert, and D. R. Roop. 1985. Organization of of mouse and human keratin 18: a component of intermediate
a type I keratin gene. J. Biol. Chem. 260:5867-5870. filaments expressed prior to implantation. Differentiation
35. Lazarides, E. 1982. Intermediate filaments: a chemically heter- 33:61-68.
ogeneous, developmentally regulated class of proteins. Annu. 57. Peabody, D. S., and P. Berg. 1986. Termination-reinitiation
Rev. Biochem. 51:219-250. occurs in the translation of mammalian cell mRNAs. Mol. Cell.
36. Lewis, S. A., and N. J. Cowan. 1986. Anomalous placement of Biol. 6:2695-2703.
introns in a member of the intermediate filament multigene 58. Peabody, D. S., S. Subramani, and P. Berg. 1986. Effect of
family: an evolutionary conundrum. Mol. Cell. Biol. upstream reading frames on translation efficiency in simian virus
6:1529-1534. 40 recombinants. Mol. Cell. Biol. 6:2704-2711.
37. Linial, M. 1987. Creation of a processed pseudogene by retro- 59. Powell, B. C., G. R. Cam, M. J. Fietz, and G. E. Rogers. 1986.
viral infection. Cell 49:93-102. Clustered arrangement of keratin intermediate filament genes.
38. Luthman, H., and G. Magnusson. 1983. High efficiency polyoma Proc. Natl. Acad. Sci. USA 83:5048-5052.
DNA transfection of chloroquine treated cells. Nucleic Acids 60. Quax, W., W. V. Egberts, W. Hendriks, Y. Quax-Jeuken, and H.
Res. 11:1295-1308. Bloemendal. 1983. The structure of the vimentin gene. Cell
39. Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular 35:215-223.
cloning: a laboratory manual. Cold Spring Harbor Laboratory, 61. Quax, W., L. van den Broek, W. V. Egberts, F. Ramaekers, and
Cold Spring Harbor, N.Y. H. Bloemendal. 1985. Characterization of the hamster desmin
40. Maniatis, T., S. Goodbourn, and J. A. Fischer. 1987. Regulation gene: expression and formation of desmin filaments in nonmus-
of inducible and tissue-specific gene expression. Science cle cells after gene transfer. Cell 43:327-338.
236:1237-1245. 62. RayChaudhury, A., D. Marchuk, M. Lindhurst, and E. Fuchs.
41. Marchuk, D., S. McCrohon, and E. Fuchs. 1984. Remarkable 1986. Three tightly linked genes encoding human type I kera-
conservation of structure among intermediate filament genes. tins: conservation of sequence in the 5'-untranslated leader and
Cell 39:491-498. 5' upstream regions of co-expressed keratin genes. Mol. Cell.
42. Marchuk, D., S. McCrohon, and E. Fuchs. 1985. Complete Biol. 6:539-548.You can also read