Addition of H19 'Loss of Methylation Testing' for Beckwith-Wiedemann Syndrome (BWS) Increases the Diagnostic Yield
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
Journal of Molecular Diagnostics, Vol. 12, No. 5, September 2010
Copyright © American Society for Investigative Pathology
and the Association for Molecular Pathology
DOI: 10.2353/jmoldx.2010.100005
Addition of H19 ‘Loss of Methylation Testing’ for
Beckwith-Wiedemann Syndrome (BWS) Increases
the Diagnostic Yield
Jochen K. Lennerz,*† Robert J. Timmerman,‡ Beckwith-Wiedemann Syndrome (BWS) is a congenital
Dorothy K. Grange,§ Michael R. DeBaun,¶ overgrowth disorder with a predisposition for develop-
Andrew P. Feinberg,储 and Barbara A. Zehnbauer** ment of aggressive embryonal-type tumors, particularly
Wilms’ tumor.1– 6 BWS is primarily a clinical diagnosis and
From the Department of Pathology,* Massachusetts General
relies on the hallmark features, including macroglossia,
Hospital/Harvard Medical School, Boston, Massachusetts; the
Departments of Pathology and Immunology,† and Pediatrics,
facial dysmorphology, visceromegaly, and macrosomia
Divisions of Genetics and Genomic Medicine,§ and Hematology-
at birth (⬎90th percentile), abdominal wall defects, neo-
Oncology,¶ Washington University School of Medicine, St. Louis, natal hypoglycemia, and ear creases or pits.2,7–11 Accu-
Missouri; the Molecular Diagnostic Laboratory,‡ Mercy Medical rate diagnosis is critical to establish effective tumor sur-
Center, Des Moines, Iowa; Johns Hopkins Medical Institutions,储 veillance via serial abdominal ultrasounds and serum
Baltimore, Maryland; and the Laboratory Practice Evaluation ␣-fetoprotein monitoring throughout childhood.2,12 Mo-
and Genomics Branch,** Centers for Disease Control and lecularly, BWS is a prototypical imprinting disorder with
Prevention (CDC), Division of Laboratory Systems, Atlanta, unequal gene expression at 11p15.5 (Figure 1, A–D).7,13–38
Georgia In addition to genetic counseling and screening in siblings,
the clinical utility of molecular confirmation is twofold. The
BWS phenotype is highly variable,2,10,11,39,40 visceral
manifestations may not be obvious at birth,10,40 facial
Beckwith-Wiedemann syndrome (BWS) is a clinical features normalize across childhood,2,40 and hemihyper-
diagnosis; however , molecular confirmation via abnor- plasia may be the only presenting feature.6,41– 45 In these
mal methylation of DMR2(LIT1) and/or DMR1(H19) has atypical or ‘variant’ cases with a milder phenotype, labo-
clinical utility due to epigenotype-tumor association. ratory confirmation may be helpful.2,6,43,46 Second, an
Despite the strong link between H19 hypermethylation epigenotype-phenotype correlation not only with specific
and tumor risk, several diagnostic laboratories only test birth defects7,47 but also with tumor-risk2,7,12,22–23,45– 46
for hypomethylation of LIT1. We assessed the added has been shown.
diagnostic value of combined LIT1 and H19 testing in a The BWS critical region at 11p15.5 can be divided into
large series of referred samples from 1298 patients, at least two functionally independent imprinting domains:
including 53 well-characterized patients from the St. a centromeric domain, DMR2 (TSSC3-KCNQ1) and a
Louis Children’s Hospital BWS-Registry (validation sam- telomeric domain, DMR1 (INS-H19).14 Normally, at least
ples) and 1245 consecutive nationwide referrals (prac- two imprinting control regions [ICR, also referred to as
tice samples). Methylation-sensitive enzymatic diges- differentially methylated regions (DMR)] regulate differ-
tion with Southern hybridization assessed loss of ential expression.12,13,48 –51 In BWS, abnormal imprinting
normal imprinting. In the validation group, abnormal may result from a variety of defects including abnormal
LIT1 hypomethylation was detected in 60% (32/52) of methylation (⬃55%), paternal segmental uniparental dis-
patients but LIT1/H19-combined testing was abnormal omy (UPD, ⬃20%), mutations/microdeletions (⬍5%), and
in 68% (36/53); sensitivity in the practice setting dem-
translocations of the maternal gene (⬍2%).22–23,51–53
onstrated 27% (342/1245) abnormal LIT1 and 32% (404/
With this heterogeneity of molecular aberrations, BWS
1245) abnormal LIT1/H19-combined. In addition, H19
poses significant challenges to the design and valida-
methylation was abnormal in 7% of LIT1-normal pa-
tients. We observed absence of uniparental disomy
(UPD) in 27% of combined LIT1/H19-abnormal sam- Supported by National Institutes of Health grant CA54358 (to A.P.F.).
ples, diagnostic of multilocus methylation abnormali- Accepted for publication March 1, 2010.
ties; in contrast to studies implicating that combined Preliminary parts of this study have been presented at the Association
LIT1/H19 abnormalities are diagnostic of UPD. The of Molecular Pathology meeting in 2002 and 2009.
overall low detection rate, even in validated patient D.K.G. has been a consultant for Biomarin Pharmaceutical, Inc.
samples and despite characterization of both loci and Address reprint requests to Barbara A. Zehnbauer, Ph.D., FACMG,
UPD status, emphasizes the importance of clinical diag- Centers for Disease Control and Prevention, Chief, Laboratory Practice
nosis in BWS. (J Mol Diagn 2010, 12:576 –588; DOI: Evaluation and Genomics Branch, 1600 Clifton Road NE, Mail Stop G23,
10.2353/jmoldx.2010.100005) Atlanta, GA 30329. E-mail: bzehnbauer@cdc.gov.
576H19 Testing Improves Diagnosis of BWS 577
JMD September 2010, Vol. 12, No. 5
Figure 1. Normal and abnormal methylation in Beckwith-Wiedemann syndrome. A: Location of markers used for UPD-analysis (filled circles, practice group;
open circles, validation group). B: Chromosomal region 11p15.5 involved in BWS-associated genomic imprinting defects. With the exception of IGF2 and LIT1,
all imprinted genes are expressed (open boxes) from the maternal allele (arrows in transcription direction); silenced genes, black boxes. C: The region can be
divided into a centromeric and telomeric domain. Mitsuya et al16 described the existence of a long QT intronic transcript (LIT1) within KCNQ1OT1 that was
transcribed in antisense orientation and is referred to as KCNQ1 overlapping transcript 1 (KCNQ1OT1). The assay targets a NotI site in the CpG island within intron
10 of the KCNQ1 gene and at the 5⬘ end of KCNQ1OT1 known as DMR217,18 (synonyms are IC2, ICR2, BWSIC2 and KvDMR1). H19 is also known as BWS, and
the assay targets a SmaI site 5 kb upstream of the H19 promoter between exon 3 and 4 of IGF2 known as DMR119 (synonyms are: IC1, ICR1, BWSIC1, H19DMR,
and CTCF binding region). Because DMR2 can refer to a centromeric17,20 or telomeric19 methylation center, in the diagnostic setting LIT1/H19 are the preferred
terms. Other differentially methylated regions (DMR) upstream of H19, not assayed here, are depicted (DMR0 located 5⬘ of the main IGF2 promoter,20 and DMR2
located between intron 7 and 9 of IGF219); composed after previous studies.7,13,22–38 D: Validation of cloned probe DNA fragments in a known BWS-UPD case.
There are no methylated LIT1 and no unmethylated H19 bands; methylation index (MI) ⫽ top band/sum approximates 0 and 1, respectively. NL, nonaffected
individual; BWS, clinical diagnosis of BWS.
tion of diagnostic assays. Typically, molecular defects type correlation of tumor risk associated with H19 hyper-
are identified in ⬃2/3 of clinically characterized BWS methylation status (inverse of IGF2), it is surprising that
patients reported in research studies.7,22–25,54 Several several clinical molecular laboratories only evaluate the
studies examined molecular detection in milder pheno- hypomethylation status of LIT1 (GeneTests, http://www.
types42,43,45,46; however, detection rates in patients re- ncbi.nlm.nih.gov/sites/GeneTests/?db⫽GeneTests, Copy-
ferred for molecular BWS testing (eg, rule out BWS) is to right 1993–2010, University of Washington, Seattle, last ac-
our knowledge unexplored. Thus, detection rates cited in cessed Feb 23, 2010). A negative test result may lead to a
reviews of BWS literature frequently refer to clinically false diagnosis without recognition of the appropriate
well-characterized BWS patients derived from research- clinical context. Our main study objective was to review
type settings.2,11,43,46,55,56 To systematically address BWS test performance in the clinical practice setting, by
BWS laboratory test performance, this study assessed comparing the diagnostic yield of LIT1 testing alone com-
molecular detection rates in the practice setting of a pared with combined LIT1/H19 assessment.
clinical molecular genetics laboratory. Furthermore, we have previously provided evidence
One successful molecular diagnostic approach is de- for a potentially inadequate classification of UPD in
tection of aberrant methylation of DMR2(LIT1) and/or BWS.7 In general, it is believed that combined LIT1/H19-
DMR1(H19) via loss of imprinting (LOI) analysis.7,22–25 In methylation abnormalities are diagnostic of UPD. While
the diagnostic setting we refer to these loci as LIT1 and this may be the case in the majority of patients, we have
H19 (Figure 1).7,13–38 The methylation status of these two previously demonstrated LOI throughout the entire BWS-
functionally independent loci normally occurs in an in- domains in the absence of UPD.7 Such multilocus meth-
verse fashion (Figure 1C)7,13–38 and has been shown to ylation abnormalities/mutations require exclusion of UPD
be representative of regulatory control by the two major via combined testing of patient and parental DNA. Again,
methylation domains.13,26,49 –51,57–58 LIT1 is normally ex- reports on frequencies in the practice setting are lacking.
pressed only from the paternally inherited gene copy, Therefore, an additional objective of the study is to de-
and H19 is normally expressed only from the maternally termine the ratio of UPD and non-UPD in the practice
inherited gene copy. Despite a high epigenotype-pheno- samples with combined LIT1/H19 abnormalities.578 Lennerz et al
JMD September 2010, Vol. 12, No. 5
To answer these questions, we use our experience
from analysis of more than 1000 nationwide referrals for
molecular testing received over an 8-year period and
comparison with data from patients with BWS from the St.
Louis Children’s Hospital–Washington University School
of Medicine BWS-registry. Our data emphasize the im-
portance of combined LIT1/H19 assessment as well as
availability of parental samples for UPD assessment
when indicated. A low sensitivity of detection by molec-
ular assays underscores the incomplete understanding
of BWS disease biology and highlights the essential role
of clinical diagnosis. The genomic Southern-based data-
set presented here forms a large-scale reference point
for new diagnostic approaches.
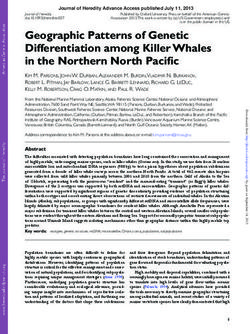
Materials and Methods Figure 2. Threshold determination in BWS via methylation hybridization. A:
Examples of autoradiographs from the validation group for the two loci.
Absence of maternal methylation pattern is more obvious and complete in
Patient Samples LIT1 when compared with the mostly incomplete hypermethylation of H19.
Methylation index (MI) ⫽ top band/sum (see Materials and Methods) pro-
This study includes analysis of a total of 1461 de-identi- vided below. B: Based on the methylation indices for the validation group,
fied patients comprising 139 samples used for methyl- diagnostic thresholds for the diagnosis of BWS were determined as ⬍0.38
(LIT1) and ⬎0.65 (H19), corresponding to mean control ⫾ 2 (details see
ation threshold determination, 53 patients in the valida- methods; P values provided in Table 3). NL, nonaffected individual; BWS,
tion group, and 1269 consecutive referred samples in the clinical diagnosis of BWS.
practice group. In this report, we focus on detection rates
and analytical aspects pertinent to molecular diagnostics
and do not include epidemiological data, tumor inci- sults) include 149 ‘in-house’ patients (St. Louis Children’s
dence, follow-up, epigenotype-phenotype correlation, or Hospital, Barnes-Jewish Hospital and affiliated hospitals,
other demographic characteristics. representing 12% of 1245) and 1096 referred patients
Methylation threshold determination samples com- (88%). Referral settings of the tests in the practice group
prised two sets: replicates and controls. Replicates con- were reviewed and analyzed for detection rates.
sisted of paired normal (age-matched) and abnormal
DNA with known methylation status for H19 and LIT1.54
Four laboratory technologists assayed these samples as LIT1/H19 Methylation Assay
30 and 32 replicates for LIT1 and H19 pairs (⫻2), respec- DNA isolation was performed from peripheral blood using
tively.54 Each pair was individually digested and hybrid- the Puregene DNA Isolation Kit according to established
ized (see below); the 124 results in 4 groups are referred protocols (Qiagen Inc., Valencia, CA).
to as R30/32 (Figure 2B). In addition to these 124 sam- Plasmid probe isolation was performed using subclon-
ples, fifteen unrelated, age-matched, nonaffected sam- ing efficiency competent cells DH5 ␣ (Invitrogen # 44-0
ples were assessed for each marker and formed the 098, Carlsbad, CA) and Qiagen Plasmid Maxi Prep (#
controls set; referred to as N15 (Figure 2, A and B). 12143, Valencia, CA). Probes for H19 (CpG island) and
Control samples were derived from healthy age-matched LIT1 (EST592241) were used to isolate cloned human
controls, available from the initial phase of diagnostic DNA fragments for Southern hybridization. The plasmids
testing associated with the BWS-registry. were digested with either Pst1/Sma1 or EcoR1/Xho1 for
Validation group refers to 53 well-characterized and H19 for LIT1, respectively. Cloned Probe DNA fragments
consented patients from the BWS Registry. Aiming at (750 bp LIT1; 1kb H19) were purified/isolated from
better understanding of the natural history and genetics of plasmids using the QIAquick gel extraction system
BWS, the registry was established in 1994 at the Genetic (Qiagen) and initial probe validation (assay develop-
Epidemiology Branch of the National Cancer Institute by ment) was performed using previously tested samples
one of the authors (M.R.D.)7 and moved to the St. Louis (Figure 1D).7,13–38
Children’s Hospital and Washington University School of
Medicine in 1999 (Beckwith-Wiedemann Syndrome Regis-
try, http://www.bws.wustl.edu/, last accessed Feb 23,
Table 1. Proposed Tumor Risk Estimates for BWS and
2010). Previously presented work established diagnostic Molecular Subgroups
criteria as well as compelling epigenotype-phenotype cor-
relation in BWS, for example with respect to tumor risk7 LIT1⫹ LIT1⫺ LIT1⫺
Group Overall H19⫺ H19⫹ H19⫹ H19⫺
(Table 1).
Practice group refers to all consecutive samples from Estimated risk (%) ⬃7.5% ⬃2% 25% 35% 10%
nationwide test referrals received over an 8.5-year period Reported range (%) 4–21 1–5 25–30 35–45 10–15
(January 2001 to June 2009, n ⫽ 1269) after exclusion of Tumor risk correlates with H19-status; compiled from 2,4,6,8,10 –12,23,
inconclusive test results. These 1245 samples (see re- 24,39,40,45,59.H19 Testing Improves Diagnosis of BWS 579
JMD September 2010, Vol. 12, No. 5
Table 2. PCR Primers (UPD Analysis)
Marker Repeat Primer sequence Product Size (bp)
D11S1338 CA Forward 5⬘-GACGGTTTAACTGTATATCTAAGAC-3’ 255–268
Reverse 5⬘-TAATGCTACTTATTTGGAGTGTG-3’
D11S1760 CA Forward 5⬘-GATCTCAAGTGTTTCCCCAC-3’ 75–110
Reverse 5⬘-AAACGATGTCTGTCCACTCA-3’
D11S988 CA Forward 5⬘-CAGAAAATAGTTCAGACCACCA-3’ 112–138
Reverse 5⬘-GGGACAAGAGAAAGTTGAACA-3’
D11S1318 CA Forward 5⬘-CCCGTATGGCAACAGG-3’ 123–145
Reverse 5⬘-TGTGCATGTNCATGAGTG-3’
THO1 AATG Forward 5⬘-GTGGGCTGAAAAGCTCCCGATTAT-3’ 179–203
Reverse 5⬘-ATTCAAAGGGTATCTGGGCTCTGG-3’
D11S922 CA Forward 5⬘-GGGGCATCTTTGGCTA-3’ 88–138
Reverse 5⬘-TCCGGTTTGGTTCAGG-3’
Markers are located along 11p; see Figure 1A.
Restriction endonuclease digestion of patient samples and iii) bacteriophage lambda DNA HindIII -digested size
was performed using two double enzyme digestions of ladder (500 bp–24kb).
genomic DNA. Briefly, the DNA methylation status was Quantification of signals followed previously published
determined using a 1.8-kb Pst1 fragment of the H19 protocols.23,54,60 Briefly, signals from the autoradio-
CpG-island and a 6-kb BamHI fragment of the LIT1 CpG- graphs were scanned and quantified using scientific im-
island, which were each analyzed by digestion with the aging software (Kodak Digital Science 1D) and ImageJ
methylcytosine-sensitive restriction endonucleases Sma1 (http://rsbweb.nih.gov/ij/, last accessed Feb 23, 2010).
and Not1, respectively (New England Biolabs, Beverley, The intensities of the LIT1⫺ (6 kb and 4.2 kb) and H19
MA). To assure complete digestion, in the H19 reaction bands (1.8 kb and 0.9 kb) bands were used to calculate
Pst1 and Sma1 restriction enzymes were added first as methylation index values (MI) according to the following
10 U/g of DNA and then a second addition of 10 U/g formula: MI ⫽ intensity top band/(intensity top band ⫹
of DNA (to “boost” the reaction) after at least 1.5–2 hours intensity bottom band). Thus, the MI represents the frac-
of incubation. The Sma1 digestion was incubated over- tion of methylated signals (Figure 2A).
night at 25°C per manufacturer’s instructions. Not1 Analysis of UPD in BWS followed previously established
(10U/g DNA) and BamH1 (4U/g DNA) enzymes were methods.7,23 Briefly, multiplex PCR analysis for UPD was
added together to the LIT1 reaction, which was incubated performed when the proband’s DNA showed abnormal
for at least 2 hours at 37°C. The H19 digestion products LOI of both LIT1and H19 and samples were available
were recovered by ethanol precipitation before electro- from the affected individuals’ parents [validation group:
phoretic fragment separation. 100% versus practice group: 329/1245 samples (⬃26%)
and 28/81 combined LIT1/H19-LOI samples (⬃35%)].
Patient and parental genomic DNA was typed with a
Agarose Gel Electrophoresis and Southern panel consisting of microsatellite markers7 [Figure 1A:
Transfer of Genomic Digests validation group: D11S988, D11S1318, D11S922 (open
circles); practice group: D11S1338, D11S1760, THO1
Fragments were separated on 1.5% (LIT1) or 2.5% (H19)
(filled circles)],7,13–38 amplified with fluorescently tagged
agarose (SeaKem LE, cat. No. 50004; FMC Bioproducts,
(Cy5.5, forward) PCR primers (Table 2). Products were
Rockland, ME) and transferred to Zetaprobe nylon mem-
analyzed by capillary electrophoresis, and raw data were
branes (Bio-Rad Laboratories, Hercules, CA) by a capil-
analyzed using Genescan and Genotyper software pack-
lary transfer technique in 0.4 N NaOH. Because H19
ages (Applied Biosystems). Peak profiles of parents and
fragments are small (1.8 and 0.9 kb, see below), only the
proband are compared to determine loss of maternal
LIT1 gel (6 and 4.2 kb) was depurinated with 0.03 N HCl
alleles consistent with paternal UPD. To differentiate be-
before Southern transfer.
tween UPD and non-UPD in the setting of combined
LIT1/H19 methylation abnormalities, parental samples (or
Hybridization and Autoradiography least a maternal sample) are mandatory.
Cloned probe DNA fragments [either the 1kb Pst1/Sma1
(H19) or the EST 592241 (LIT1)] were radiolabeled using Assay Development and Validation
random priming with [␣-32P]dCTP and then incubated
with membranes for approximately 16 hours with at 65°C We assessed range, distribution, variance, average, and
or 62°C, respectively; membranes were subsequently SD within and between groups of the 2⫻ R30/32 samples
washed and subjected to autoradiography. Every exper- of the replicate set as well as in and between the control
iment in the practice group included the following refer- set (Figure 2B). Adjusted standard deviations for each
ence materials: i) Negative control DNA (unaffected indi- locus were calculated to account for the inherent analyt-
vidual), ii) Positive control DNA (ie, validation group/BWS ical imprecision (determined in the replicates) and the
registry sample or previously tested positive samples), biological variability (determined in the controls). There-580 Lennerz et al
JMD September 2010, Vol. 12, No. 5
fore we adjusted the SD according to the following for-
mula: adj ⫽ 公 [(R30/32)2 ⫹ (N15)2] adapted after61
⫽ SD; adj ⫽ adjusted; R30/32 ⫽ replicates;
N15 ⫽ controls.
Diagnostic threshold determination was performed us-
ing i) distribution of control and replicate samples deter-
mined here (R30/32; N15; Figure 2B), ii) previously re-
ported MI from samples in the BWS registry,7,43 and ii)
the adjusted standard deviations for H19 and LIT1. An
average MI for normal plus 2 standard deviations (H19)
and an average MI for normal minus 2 standard devi-
ations (LIT1) included all of the affected individuals
with abnormal MI (Figure 2B). Sensitivity of the assay
was determined using the validation group. ‘Diagnostic
yield’ was defined as assay sensitivity in relation to the
methylated regions assessed, specifically LIT1 alone
versus LIT1/H19 combined. We compared detection
rates in the validation group and the practice group
(Figure 3, A–C).
Analytical variation and robustness was approximated
by selection of positive control samples run at least six
times (range, 6 –16) during the last 2 years of the study
(2007–2009). MIs for each of the five samples were com-
pared as groups (n ⫽ 46 H19 and n ⫽ 54 LIT1), per
sample (repetitive measurements), and via variances ob-
tained during the threshold determination experiments.
Controls from the last 75 runs were compared similarly
and ‘new’ diagnostic thresholds were determined; ro-
bustness was assessed by comparison of these ‘new’ to
the original MI threshold values.
Statistical testing includes two-tailed t-tests with
Welch’s correction for unequal variances, when appro-
priate. Gaussian distribution was confirmed via the
D’Agostino-Pearson omnibus K2 normality test62 and
we assessed homoscedasticity (homogeneity of vari-
ances) by F and Bartlett’s tests.63,64 Statistical analysis
was performed using Prism5 (GraphPad Software Inc.,
San Diego, CA), and P values of ⬍0.05 were consid-
ered significant.
Results
The assay for LOI in BWS determines the methylation Figure 3. Addition of H19 increases diagnostic yield. A: In the validation
status of H19 and LIT1 by digestion with methylation group, the addition of H19-testing allows detection of four additional patients
sensitive restriction enzymes, followed by genomic in the LIT1-normal group, which corresponds to ⬃8% increased detection.
B: In the practice group, the addition of H19 allows identification of 62
Southern hybridization with DMR-specific probes. Nonaf- additional patients, missed by LIT1 testing alone. This corresponds to an
fected individuals (NL) show equally intense probe sig- overall ⬃5% increased detection rate. C: Southern blot examples from the
nals (Figure 1D, NL)7,13–38 equivalent to a MI of 0.5 practice group illustrate the four possible band combinations and are shown as
LIT1 and H19 combinations (although run as separate tests). The diagnostic
(expressed as methylated fraction over total band inten- interpretation (Interpr.) is provided under the methylation index (MI) ⫽ top
sities). In contrast, an abnormal methylation pattern (BWS) band/sum (see Materials and Methods). ⫹, abnormal; ⫺, normal methylation.
is characterized by absence of the maternal band at either
one or both loci (Figure 1D, BWS).7,13–38 Probe validation
using previously characterized samples showed appro- bution of MI (examples in Figure 2A) in this group, con-
priate performance. An example of a BWS patient with sisting of replicates and controls (Figure 2B), was used to
known UPD, confirmed by microsatellite analysis (not determine the separation between normal and abnormal
shown), illustrates the methylation pattern at LIT1 and MI. The range of MI in the replicates was: LIT1-NL 0.4 –
H19 with inversely related MI values that approach 0 and 0.59 versus LIT1-BWS 0 – 0.04 and H19-NL 0.48 – 0.64
1, respectively (Figure 1D, BWS). versus H19-BWS 0.76 – 0.94. The range of MI in the con-
Diagnostic threshold determination for LIT1 and H19 trols was LIT1: 0.46⫺0.53 versus H19: 0.45– 0.63. Fre-
followed previously published protocols.7,22,23,54 Distri- quency plots (distribution) and mean MI ⫾ for these sixH19 Testing Improves Diagnosis of BWS 581
JMD September 2010, Vol. 12, No. 5
Table 3. Statistical Summary of Methylation Index initially obtained values (t-test; Table 3), with the excep-
Comparisons tion of LIT1⫹ samples that showed more complete hy-
Comparison
pomethylation averages (P ⬍ 0.001); ‘newly’ obtained
between LIT1 H19 threshold values are almost identical to those determined
at the beginning of the study period: ⬎0.69 for H19 and
NL (Replicates) vs. P ⬍ 0.0001 P ⬍ 0.0001 ⬍0.386 for LIT1. These results confirm a substantial in-
BWS CI: 0.439–0.477 CI: 0.285–0.332
NL (Controls) vs. P ⬍ 0.0001 P ⬍ 0.0001 terrun and overall robustness of the assay.
BWS CI: 0.466–0.489 CI: 0.33–0.389
NL (TD) vs. P ⫽ 0.066 P ⫽ 0.387
‘new’ NL CI: 0.0009–0.028 CI: 0.026–0.066 Validation Group
BWS (TD) vs. P ⬍ 0.0001 P ⫽ 0.55
‘new’ BWS CI: 0.004⫺0.01 CI: 0.057–0.11 All patients in the validation group were evaluated by
P values (t-tests) and 95% confidence interval of the difference two of the authors (M.R.D., A.P.F.) at the annual BWS
between selected groups (CI); see results for details. NL indicates Registry meeting in St. Louis and fulfilled previously
nonaffected individual; BWS, clinical diagnosis of Beckwith-Wiedemann published criteria.7 Briefly, each patient had a clinical
Syndrome; Replicates, 30(LIT1) and 32(H19) replicate pairs assessed
by four laboratory technologists; Controls, 15 unrelated, unaffected diagnosis of BWS (made by a physician) with at least two
samples; TD, threshold determination; ‘new,’ controls of the 75 most of the five most common features associated with
recent runs in the practice setting.
BWS,7,9,10 including macroglossia, birth weight and
length ⬎90th percentile, hypoglycemia in the first month
of life, ear creases or ear pits, and midline abdominal wall
groups is provided in Figure 2B. The differences in MI
defects (omphalocele, diastasis recti, or umbilical her-
between NL and BWS were statistically highly significant:
nia). For clinical characteristics of the validation group we
LIT1 R30 (n ⫽ 30 NL versus 30 BWS, P ⬍ 0.0001, t ⫽
refer to our previously published case-cohort study.7 In
49.07) and H19 R32 (n ⫽ 32 NL versus 32 BWS, P ⬍
addition to the well-established phenotypic traits, we spe-
0.0001, t ⫽ 26.26, two-tailed t-tests). Similarly, the differ-
cifically chose this subset of 53 patient samples because
ences between controls and BWS samples were statisti-
they comprised the original validation set for clinical test-
cally significant (H19 N15: P ⬍ 0.0001, t ⫽ 23.57; LIT1
ing.54 We determined the sensitivity of the assay by using
N15: P ⬍ 0.0001, t ⫽ 68.54, two-tailed t-tests); for details
the MI thresholds in the validation group (Figure 3A).
see Table 3. To accurately estimate total variability, stan-
Abnormal methylation for LIT1 was found in 32 of the 53
dard deviations of replicates and controls were adapted
patients (LIT1 detection rate 60%). Testing for H19 in the
(see Materials and Methods) and the diagnostic threshold
validation group identified 10 patients with abnormal
set to ⫾ 2 of the normal MI for each locus. MI ⬎0.65 for
methylation, four of whom would have been labeled ‘neg-
H19 and ⬍0.38 for LIT1 are abnormal and are referred to
ative’ based on LIT1 testing alone. Thus, H19 testing
as H19⫹ and LIT1⫹ (Figure 2B). These thresholds com-
increases the overall sensitivity by 8% (combined LIT1
pare to those reported in the literature.7,22–25
and H19 sensitivity 68%). Interestingly, the proportions of
From an analytical perspective we determined normal
H19⫹ samples in the LIT1⫹ and LIT1-normal groups were
distribution of MI (P range: 0.21– 0.17; K2) and found
equal (10%).
equal variances within two (N30/32 versus BWS30/32; P
The six additional cases in this LIT1⫹ group repre-
range: 0.73– 0.92, F test) or multiple groups (N30/32,
sented paternal UPD inheritance [6/53; confirmed via
N15, BWS; P range: 0.64 – 0.82 Bartlett’s test). Unexpect-
parental samples (see below)] and 19% of the LIT1-
edly, we noticed significantly different (analytical) vari-
normal patients had H19 abnormalities (4/21). Overall in
ances when comparing between loci (LIT1 versus H19,
this population, the occurrence of a single locus with
P ⫽ 0.0064 NL; P ⫽ 0.002– 0.004 F tests). The frequency
aberrant methylation was found in 26 (49%) and 4 (7.5%)
plots of abnormally methylated LIT1 and H19 (black bars
patients for LIT1 and H19, respectively. These propor-
in Figure 2B; bin width 0.05) confirm this and illustrate i)
tions as well as the ⬃11% UPD are in concordance with
almost complete demethylation of LIT1 versus ii) a higher
published findings in well-characterized patients with
proportion of incomplete hypermethylation of H19; these
BWS7.22–25,54 In the validation group, we did not detect
results have not only analytical but biological implications
combined H19/LIT1-LOI in the absence of UPD, consis-
(see discussion).
tent with previous reports.7,24,26,69
Control DNA from cultured cells cannot be used as
reference material because the methylation status is not
maintained during cell culture conditions,7,27,65– 68 thus Practice Group
we used separate reference materials (see Materials and
Methods). We tested the robustness of the assay by First, we determined the technical/analytical sensitivity in
comparison of variances of positive controls, taken from practice by review of the inconclusive results (H19 n ⫽
the last 2 years of the study period (P range: 0.07– 0.21 17; LIT1 n ⫽ 24) and found that insufficient quality or
Bartlett’s test), as well as the simulated determination of purification of DNA was one of the major reasons for
‘new’ methylation thresholds. When MIs were obtained these results (⬃46% of inconclusive results). Other fac-
from positive and negative controls in each of the 75 most tors contributing to inconclusive results may be sample
recent runs [LIT1⫺: 0.462 ⫾ 0.038; LIT1⫹: 0.011 ⫾ 0.002; interference (ie, inhibiting endonuclease activity) or an
H19⫺: 0.518 ⫾ 0.086; H19⫹: 0.833 ⫾ 0.231] there was no undetermined factor. Based on the number of inconclu-
significant deviation of these threshold values from the sive test results from the practice group, the analytical582 Lennerz et al
JMD September 2010, Vol. 12, No. 5
sensitivity was determined to be 98.1% for LIT1 (1245/ Table 4. Molecular Detection Rates by Referral Setting
1269) and 98.6% for H19 (1252/1269), which further con-
Practice
firms the overall robustness of the assay. After exclusion group ‘Geneticists’ ‘Non-Geneticists’ ⌺
of seven samples without LIT1 results, the practice group
and final analysis was performed on the remaining 1245 Abnormality n ⫽ 313 n ⫽ 79 n ⫽ 392
patients’ samples. detected
No molecular n ⫽ 658 n ⫽ 163 n ⫽ 821
Diagnostic yield of LIT1 testing and the addition of H19 abnormality
testing were also determined in the practice group (Fig- Detection rate 32.2% 32.6% 32.3%
ure 3B); examples of Southern blots and MI for each of
For explanation of ‘geneticists’ and ‘non-geneticists’ see Results and
the four diagnostic groups are provided in Figure 3C. Of Discussion.
the 1245 patients we found abnormal methylation for LIT1
in 342. This LIT1-based detection rate of 27% is markedly
lower than in the validation group (60%). Additional test- settings were assigned to the ‘geneticist’ group. Samples
ing for H19 identified 143 abnormal patients, 62 of whom submitted from private practice physicians or pediatri-
would have been labeled ‘negative’ based on LIT1 testing cians, community hospitals, or other centers without des-
alone. Thus, addition of H19 testing increased the overall ignated genetics clinics/divisions were assigned to the
sensitivity by ⬃5% (overall sensitivity: 32%), correspond- ‘non-geneticist’ group (Table 4). Comparison of detection
ing to a relative increase of 15.6% (5/32). In total, 7% of rates showed no significant difference of molecularly ab-
the LIT1 normal patients had H19 abnormalities (62/903) normal samples referred from the ‘geneticist’ group (313/
and the 81 additional cases in the LIT1⫹ group represent 971) versus the ‘non-geneticist’ group (79/242; 2 ⫽
combined methylation abnormalities in H19 and LIT1 0.015; Fisher’s exact test P ⫽ 0.94).
(6.6%). Combined methylation abnormalities of H19 and
LIT1 either represent UPD or an imprinting disturbance
extending throughout the 11p15 imprinted domains.7 Discussion
Overall parental samples were available in ⬃26% of the
practice group (329/1245) and in 28 (35%) of the 81 We report the molecular detection rate for BWS in the
combined H19/LIT1-abnormal samples. practice setting with 1245 patients. LIT1 methylation test-
In combined H19/LIT1-abnormal samples, assessment ing alone was abnormal in 27% of samples, whereas
for UPD was performed whenever parental samples were combined assessment of H19 and LIT1 was abnormal in
available (n ⫽ 28). Lack of heterozygosity in any of the 32% of samples. Thus, 5% of children with an epigeno-
assessed alleles is considered noninformative, and we type that shows the highest tumor risk (H19) would be
excluded six cases. Absence of peaks in the proband’s missed (false-negatives) using LIT1 LOI assessment
traces that correspond to unique maternal peaks indi- alone. The sensitivity of the assay was determined in the
cates loss of maternal alleles (consistent with paternal validation group of 53 well-characterized patients with
UPD), which we confirmed in 16 of the informative cases BWS and the result was abnormal in ⬃60% of cases with
(73%). In 12 of these 16 cases consistent with paternal LIT1 testing alone versus ⬃68% of cases with combined
UPD, the proband’s sample showed minimal evidence of H19/LIT1 assessment. These findings highlight the rela-
maternal-specific peaks (⬍30% of the peak-height in tively low detection rate in both settings and thereby
comparison with paternal alleles70), which was inter- underscore the significant role of clinical diagnosis in
preted as mosaicism (see Discussion). The remaining six patients with BWS (Figure 4, A and B). Furthermore, in the
informative cases showed either identical biparental al- group of combined abnormal methylation status (H19/
lelic contribution (four cases) or minimal peak-height dif- LIT1), we found UPD in 73% whereas UPD was excluded
ferences with ⬍20% skewing7,70 (two cases). These find- in 27%. Thereby we verify the existence of multilocus
ings exclude UPD in six of 22 informative cases (27%; methylation mutations in the molecular diagnostic prac-
this number excludes three previously reported cases7). tice setting, which underscores the importance of as-
Furthermore, cases that harbor LIT1 and H19 methylation sessment of UPD in addition to epigenetic testing in BWS.
mutations in the absence of UPD are diagnostic of mul- Clinically, BWS can be separated into three major sub-
tilocus methylation abnormalities that extend through groups, familial, sporadic, and those with chromosome
both domains at 11p15.5. abnormalities.26,71 To account for differences in these
Referral settings were analyzed to identify differences subgroups (eg, higher rate of CDKN1C mutations in fa-
in detection rates. For 1213/1245 (97.4%) of the samples milial cases), and to decrease limitations associated with
the referral setting was available; 32 samples had to be individual diagnostic methods, multimodal algorithms
excluded from this analysis due to lack of specific infor- have been proposed.2,17 Knowing the limitations of mo-
mation. First we determined the proportion of molecularly lecular tests in these algorithms becomes paramount in
abnormal samples in the subgroup where the referral the controversial context of prenatal testing or increased
setting was known (392/1213) versus that observed in the BWS rates in children conceived via some assisted re-
entire practice group (404/1245) and found no statisti- productive technologies,47,76 – 80 especially because the
cally discernable difference (Fisher’s exact test, P ⫽ phenotype cannot be ascertained prenatally. There is a
0.97). Next, we separated the referral settings into two need to diagnose the syndrome earlier, afford better
groups: samples submitted from geneticists and desig- counseling to the parents, and improve neonatal care of
nated genetics clinics in academic and non-academic patients.81 However, in the absence of better molecularH19 Testing Improves Diagnosis of BWS 583
JMD September 2010, Vol. 12, No. 5
that have both maternal and paternal alleles mixed with
lymphocytes that are missing the maternal allele. Thus,
mosaicism refers to the presence of paternal isodisomy
(affecting at least a segment of 11p) in addition to a
normal cell line,90 which has at least two practical impli-
cations. First, due to differential involvement of tissues,
the true rate of UPD is likely higher than that reported
here or elsewhere.11,23 Second, and importantly for ge-
netic counseling, mosaicism in BWS indicates that UPD
arises as a postzygotic event,72,90 –91 which supports the
empirically very low recurrence risk (approximating that
of probands with affected siblings)2,69,84 and offers an
explanation for the intriguing reports of monozygotic twin
pairs discordant for BWS.92–100 In contrast, the exact recur-
rence risk in non-UPD BWS is unknown,7,11 but is likely low
and cursorily mentioned as ⬍5%101 (GeneTests, http://
www.ncbi.nlm.nih.gov/sites/GeneTests/?db⫽GeneTests,
Copyright 1993–2010, University of Washington, Seattle,
last accessed Feb 23, 2010); exception CDKN1C muta-
tions (see below). From a practical standpoint, abnormal
imprinting in both domains may functionally reflect UPD
and several studies have demonstrated such correla-
tion.11,24 However, the inference that epigenetic testing is
sufficient to assess UPD has to be put in perspective of
our previous findings that 25% of cases with combined
methylation abnormalities at H19 and LIT1 are not con-
sistent with UPD by microsatellite analysis.7 Demonstra-
tion of a similar frequency (27% non-UPD in combined
abnormal cases versus 73% UPD) in a larger series of
patients suggests the existence of this molecular sub-
group within the clinical genetics practice setting. The
alterations are methylation abnormalities extending
Figure 4. Diagnostic algorithm and molecular diagnostic approach in BWS. throughout the 11p15.5 domain.7 Underlying mecha-
A: The diagnostic algorithm follows clinical (phenotypic) features and key nisms, recurrence risk, and clinical significance of mul-
examples are provided to illustrate the following interrelation: with decreas- tilocus (‘coordinated’) methylation mutations remain to be
ing clinical suspicion of BWS, the rate of molecular detection decreases;
however, clinical variability or specific settings may trigger testing and con- determined. However, LOI at multiple maternally methyl-
firm abnormal methylation in ‘mild phenotypes’ or hemihyperplasia6,41– 45 ated loci, including loci outside of 11p15.5, has led to the
(for comprehensive coverage of clinical features see Refs.7–10,59,71–75). As a
visual estimate, the detection rates in the validation and practice group are
proposal of a ‘maternal hypomethylation syndrome’ in
provided (gray background). B: Epigenetic testing and assessment of unipa- BWS.102,103 Recently multilocus methylation defects with
rental disomy (UPD) should be combined, and the required samples are simultaneous loss of paternal and maternal imprinted loci
listed. Finally, the interpretation of the test result and institution of tumor
surveillance procedures requires clinical correlation and additional diagnos- has been demonstrated in BWS72; however, we were not
tic testing (eg, cytogenetics or CDKN1C sequencing). able to review clinical features in our referred cases and
can neither support nor refute a reported trend of tumor
association in multilocus methylation defects.72 Including
understanding of the disease biology, coupled with an the phenotypic variation observed in genome-wide pa-
absence of comprehensive diagnostic assays, BWS re- ternal UPD,70,104 –106 the distinction of UPD from coordi-
mains a clinical diagnosis. Thus, currently molecular test- nate/multilocus methylation defects is more than an aca-
ing may confirm the diagnosis but cannot rule it out2,82 demic exercise and the extent of molecular abnormalities
(Figure 4B). may determine the exact phenotype in BWS. Although
Implementation of epigenetic testing has expanded the practical implications of multilocus methylation de-
our toolkit for molecular confirmation; nonetheless one of fects are uncharted, the assessment of UPD remains a
the basic tenets in laboratory testing in BWS remains the cornerstone of genetic testing and counseling. Despite
assessment of UPD with its implications for genetic coun- frequent elucidation in reviews, it is surprising that one
seling83 and recurrence risk.84 Whenever possible, pa- practical aspect of BWS testing has not been empha-
rental samples (or at least the maternal) should be ob- sized strongly enough, namely the submission of parental
tained and submitted with the proband samples for samples. Specifically, the assessment of UPD requires
epigenetic assessment. Paternal UPD in BWS is essen- microsatellite analysis that includes the proband and, at
tially always partial/segmental22,71,85– 89 and somatically least, the mother.
mosaic.22,23,89,90 This was confirmed by the presence of Detection rates are related to diagnostic threshold cut-
low-intensity maternal allele-peaks in the UPD-analysis, offs and the high-variability of the latter7,22–25,54 empha-
likely reflecting a low number of circulating lymphocytes sizes the quality requirement for well-documented in-584 Lennerz et al
JMD September 2010, Vol. 12, No. 5
house validation.54 For instance, if thresholds as reported controversial for LIT1,24,25,26,69 we have previously shown
in the published literature were used (eg, H19: 0.50), at that there is no difference in the frequency of increased H19
least seven of the fifteen normal controls would be falsely methylation when comparing BWS samples from lym-
labeled as H19-positive. Because determination of diag- phoctyes or BWS samples from fibroblasts.7 Thus, part of
nostic/MI-cut-off values also applies to newer methods the low detection rate derives from our incomplete under-
discussed below, the importance of threshold determina- standing of BWS genetics and how to transition the best
tion and assay validation cannot be overemphasized. We targets into diagnostic practice. Our data specify the
recognized the critical importance of complete restriction presence of a substantial number of LIT1-normal patients
endonuclease digestion from our prevalidation experi- with H19 abnormalities, indicating that established tar-
ence; the importance of complete capillary transfer of all gets should be used in a meaningful way.
sizes of DNA fragments during Southern blotting; and the The overall low detection rate in the practice setting
optimization and standardization of specific radiolabel- with 27% (LIT1 testing alone) and 32% (combined LIT1/
ing, hybridization, autoradiography, and scanning condi- H19 testing) goes beyond the known limitations of mo-
tions (see Materials and Methods). lecular diagnostic testing in BWS and suggests other
In addition to analytical challenges, another reason restrictions. For example, we know that the practice
H19 testing is not widely implemented may be biological. group is more heterogeneous because samples were
Epigenetic regulation of the DMR subdomains occurs submitted for various reasons (eg, “exclude BWS”) and
independently,16,22–25,28,107 and while there is at least testing was not refused based on the absence of pub-
one imprinting center in the centromeric domain, there lished BWS criteria (eg, “atypical features,” “isolated
are at least three imprinting centers13,28 –30 in the telo- hemihyperplasia”). In fact, individual ‘in house’ cases did
meric domain influencing H19/IGF2 expression (Figure not show the classic diagnostic features but had an ab-
1B).7,13,16 –38 This difference in organization may explain normal methylation pattern. Although the clinical diversity
our findings of nearly complete demethylation of LIT1 of BWS has very recently been extended to include a
(Figures 2, A and B, and 3C) whereas H19 is frequently neurobehavioral phenotype with genotype correlation,116
incompletely hypermethylated (Figures 2B and 3C). Fur- our samples were referred for testing before the descrip-
thermore, despite reciprocal H19/IGF2-locus interdepen- tion of these ‘new’ phenotypic features. In another ap-
dence23,26,108 –110 referred to as ‘CTCF enhancer com- proach to explain the substantially lower percentage of
petition model’,26,110 –112 the complex organization in the molecularly abnormal samples in the practice group, we
telomeric domain may also explain biallelic IGF2 expres- compared detection rates based on referral setting and
sion with normal H19 methylation.89,113,114 IGF2 is pre- found no difference (Table 4). The distinction of referral
sumed to be one contributing element to the BWS over- setting is inexact and therefore a weak estimate for the
growth phenotype and accordingly, distinction between influences of subspecialty expertise on detection rates.
H19-dependent and -independent IGF2 expression has Due to limited availability, we did not review reasons for
been made.22,23,26,31,110,113,114 H19-indepenent IGF2 ex- referral to a geneticist, reasons for molecular testing (for
pression has mostly been demonstrated in fibroblast cul- example, insurance coverage), or the fact that not all
tures and maintenance of methylation pattern remains patients chose to be tested, which may introduce selec-
questionable.7,27,65– 68 However, based on very few ex- tion bias. However, because the authors were not in-
ceptions demonstrated in peripheral blood23,114 one may volved in any of these decisions, ultimately selection of
argue that addition of IGF2 testing may increase the the practice group relied on clinical diagnostic judgment
diagnostic yield in BWS testing as well. We identified of the referring physicians, which fulfills the designation,
three substantial hindrances for the latter: i) There is a ‘practice setting.’
lack of polymorphic markers to determine uni- versus Although the methods applied here are considered the
biallelic expression of IGF2 (unpublished observations). ‘gold-standard’ in BWS molecular testing, some of the
ii) To date, only a single study has combined H19/LIT1 genomic Southern hybridization associated limitations
and IGF2 testing21 and no IGF2 hypermethylation was are: technical difficulty, large quantities of required input
observed when H19 imprinting was normal (⬎80 sam- DNA (⬃10 g), significant time commitment with ex-
ples). Given the identical diagnostic yield of 68%21 and tended turn-around-time, and high personnel cost. Fur-
the rather high variability of IGF2 methylation assessment thermore, each locus must be analyzed separately and
(personal communication, Dr. Andrea Riccio), IGF2 test- information about copy number cannot be reliably ob-
ing in routine diagnostics is not feasible. iii) Furthermore, tained. PCR-based methods overcome some of these
conversion of IGF2 analysis from basic science into di- limitations, and distinction of methylation status can be
agnostic practice is not straightforward because expres- achieved via sodium bisulfite modification of the genomic
sion is restricted to particular tissues (eg, in tongue, DNA before PCR (methylation of cytosine conveys resis-
kidney, placenta) and is absent in peripheral blood tance to bisulfite treatment whereas unmethylated cy-
mononuclear cells which form the source of genomic tosines are deaminated to uracil). Methylation-specific
DNA in the routine diagnostic setting.23,24,27,32–37,115 This PCR (MS-PCR) has been established in BWS diagnos-
leads to another reason for the difficulty with molecular tics117 and other variations include combined bisulfite
assessment not only of H19 but BWS in general: blood restriction assay (COBRA: bisulfite PCR⫹ restriction en-
cells are not involved in the BWS phenotype but used as zyme step),118 bisulfite pyrosequencing,119 –121 or high-
a routine surrogate for diagnostic testing. While expres- resolution melting analysis (HRM).122–124 Despite several
sion differences between blood and affected tissues is advantages of these approaches, recently, methylation-H19 Testing Improves Diagnosis of BWS 585
JMD September 2010, Vol. 12, No. 5
sensitive multiplex ligation probe analysis (MS-MLPA) 6. Tan TY, Amor DJ: Tumour surveillance in Beckwith-Wiedemann syn-
has been introduced and deemed the ‘platinum-stan- drome and hemihyperplasia: a critical review of the evidence and
suggested guidelines for local practice. J Paediatr Child Health
dard.’ MS-MLPA can detect microdeletions, microdupli- 2006, 42:486 – 490
cations, alterations in gene dosage, as well as DNA meth- 7. DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP,
ylation, including UPD.2,17,73 Notwithstanding these Feinberg AP: Epigenetic alterations of H19 and LIT1 distinguish
advantages, MS-MLPA-detection rates are similar to patients with Beckwith-Wiedemann syndrome with cancer and birth
defects. Am J Hum Genet 2002, 70:604 – 611
former standards and ⬃78% in well-characterized pa-
8. Elliott M, Bayly R, Cole T, Temple IK, Maher ER: Clinical features and
tients with BWS.125 Therefore, none of the current meth- natural history of Beckwith-Wiedemann syndrome: presentation of
ods, including those applied here, are able to replace a 74 new cases. Clin Genet 1994, 46:168 –174
multimodal approach including detection of transloca- 9. Elliott M, Maher ER: Beckwith-Wiedemann syndrome. J Med Genet
tions/inversion via karyotyping and CDKN1C alterations 1994, 31:560 –564
10. Pettenati MJ, Haines JL, Higgins RR, Wappner RS, Palmer CG,
by DNA sequencing.2 Cytogenetic abnormalities are ob-
Weaver DD: Wiedemann-Beckwith syndrome: presentation of clini-
served in ⬃1% (FISH: ⬃2%) and CDKN1C mutations cal and cytogenetic data on 22 new cases and review of the litera-
(occurring independent of H19/LIT122,38,46,52,69,126) have ture. Hum Genet 1986, 74:143–154
been described in 1–3% of sporadic and 5–10% of famil- 11. Weksberg R, Shuman C, Smith AC: Beckwith-Wiedemann syn-
ial BWS.11,22,53 We are aware of at least three families drome. Am J Med Genet C Semin Med Genet 2005, 137C:12–23
12. Rump P, Zeegers MP, van Essen AJ: Tumor risk in Beckwith-Wiede-
(⬃2%) in the 149 ‘in house’ patients with CDKN1C muta- mann syndrome: a review and meta-analysis. Am J Med Genet A
tions. On the other hand, the recurrent theme in BWS 2005, 136:95–104
testing is that ⬃20% of well-characterized patients with 13. Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER:
BWS do not have currently-known molecular abnormali- Imprinting mutations in the Beckwith-Wiedemann syndrome sug-
gested by altered imprinting pattern in the IGF2–H19 domain. Hum
ties at 11p15.5. Consequently, epigenetic alterations
Mol Genet 1995, 4:2379 –2385
other than methylation have been proposed,26,113 and 14. Delaval K, Wagschal A, Feil R: Epigenetic deregulation of imprinting
recently two new genes, NALP2127 and ZFP57,74 located in congenital diseases of aberrant growth. Bioessays 2006, 28:
on chromosomes 19q13.42 and 6p22.1, respectively, 453– 459
have been linked to BWS. Interestingly, these genetic 15. Delaval K, Feil R: Epigenetic regulation of mammalian genomic
imprinting. Curr Opin Genet Dev 2004, 14:188 –195
abnormalities outside the 11p15.5 domains were re-
16. Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H,
stricted to individuals with LIT1 abnormalities,128,129 ar- Yoshida MA, Niikawa N, Feinberg AP, Oshimura M: LIT1, an im-
guing that multilocus methylation abnormalities extend printed antisense RNA in the human KvLQT1 locus identified by
beyond 11p15.5 but also that current testing strategies screening for differentially expressed transcripts using monochro-
capture these subgroups. Nonetheless, it remains to be mosomal hybrids. Hum Mol Genet 1999, 8:1209 –1217
17. Scott RH, Douglas J, Baskcomb L, Nygren AO, Birch JM, Cole TR,
determined to what extent new approaches may increase Cormier-Daire V, Eastwood DM, Garcia-Minaur S, Lupunzina P, Tat-
diagnostic sensitivity and how to explore the challenging ton-Brown K, Bliek J, Maher ER, Rahman N: Methylation-specific
group of molecularly negative patients with BWS. multiplex ligation-dependent probe amplification (MS-MLPA) ro-
bustly detects and distinguishes 11p15 abnormalities associated
with overgrowth and growth retardation. J Med Genet 2008,
45:106 –113
Acknowledgments 18. Coffee AL, Kuehl TJ, Willis S, Sulak PJ: Oral contraceptives and
premenstrual symptoms: comparison of a 21/7 and extended regi-
We thank the patients and families who have participated men. Am J Obstet Gynecol 2006, 195:1311–1319
in the BWS-registry and our colleagues for referral of 19. Weber M, Hagege H, Murrell A, Brunel C, Reik W, Cathala G, Forne
analytical samples to our laboratory. Finally, we are very T: Genomic imprinting controls matrix attachment regions in the Igf2
grateful for the expertise of Emily Niemitz who helped in gene. Mol Cell Biol 2003, 23:8953– 8959
20. Coffee B, Muralidharan K, Highsmith WE Jr, Lapunzina P, Warren
early phases of the assay development and acknowl- ST: Molecular diagnosis of Beckwith-Wiedemann syndrome using
edge Beverly Gibson, Jack Shields, Jeanne Anderson, quantitative methylation-sensitive polymerase chain reaction. Genet
Stephanie Adelsberger, Brooke Stroup, and Holly Simon for Med 2006, 8:628 – 634
excellent technical assistance during the study period. 21. Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, Silengo MC,
Spreafico F, Perotti D, De Crescenzo A, Sparago A, Cerrato F, Riccio
A: Distinct methylation changes at the IGF2–H19 locus in congenital
growth disorders and cancer. PLoS One 2008, 3:e1849
References 22. Bliek J, Maas SM, Ruijter JM, Hennekam RC, Alders M, Westerveld
A, Mannens MM: Increased tumour risk for BWS patients correlates
1. Beckwith JB: Extreme cytomegaly of the adrenal cortex, omphalo- with aberrant H19 and not KCNQ1OT1 methylation: occurrence of
cele, hyperplasia of kidneys and pancreas, and Leydig-cell KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol
hyperplasia: another syndrome? Abstract, Western Society for Pe- Genet 2001, 10:467– 476
diatric Research 1963 23. Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C,
2. Weksberg R, Shuman C, Beckwith JB: Beckwith-Wiedemann syn- Steele L, Cameron J, Smith A, Ambus I, Li M, Ray PN, Sadowski P,
drome. Eur J Hum Genet 2010, 18:8 –14 Squire J: Tumor development in the Beckwith-Wiedemann syn-
3. Wiedemann HR: [Familial Malformation Complex with Umbilical Her- drome is associated with a variety of constitutional molecular 11p15
nia and Macroglossia–a “New Syndrome?”] J Genet Hum 1964, alterations including imprinting defects of KCNQ1OT1. Hum Mol
13:223–232 Genet 2001, 10:2989 –3000
4. DeBaun MR, Tucker MA: Risk of cancer during the first four years of 24. Gaston V, Le Bouc Y, Soupre V, Burglen L, Donadieu J, Oro H,
life in children from The Beckwith-Wiedemann Syndrome Registry. Audry G, Vazquez MP, Gicquel C: Analysis of the methylation status
J Pediatr 1998, 132:398 – 400 of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis
5. DeBaun MR, Siegel MJ, Choyke PL: Nephromegaly in infancy and and prognosis of Beckwith-Wiedemann syndrome. Eur J Hum Genet
early childhood: a risk factor for Wilms tumor in Beckwith-Wiede- 2001, 9:409 – 418
mann syndrome. J Pediatr 1998, 132:401– 404 25. Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S,586 Lennerz et al
JMD September 2010, Vol. 12, No. 5
Oshimura M, Feinberg AP: Loss of imprinting of a paternally ex- and occurs following assisted reproductive technologies, Am J Med
pressed transcript, with antisense orientation to KVLQT1, occurs Genet A 2006, 140:1497–1503
frequently in Beckwith-Wiedemann syndrome and is independent of 45. Wiedemann HR: Tumors and hemihypertrophy associated with
insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 1999, Wiedemann-Beckwith syndrome. Eur J Pediatr 1983, 141:129
96:5203–5208 46. Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R,
26. Maher ER, Reik W: Beckwith-Wiedemann syndrome: imprinting in Bowdin SC, Riccio A, Sebastio G, Bliek J, Schofield PN, Reik W,
clusters revisited. J Clin Invest 2000, 105:247–252 Macdonald F, Maher ER: Molecular subtypes and phenotypic ex-
27. Reik W, Constancia M, Dean W, Davies K, Bowden L, Murrell A, Feil pression of Beckwith-Wiedemann syndrome. Eur J Hum Genet
R, Walter J, Kelsey G: Igf2 imprinting in development and disease. 2005, 13:1025–1032
Int J Dev Biol 2000, 44:145–150 47. DeBaun MR, Niemitz EL, Feinberg AP: Association of in vitro fertili-
28. Murrell A, Heeson S, Cooper WN, Douglas E, Apostolidou S, Moore zation with Beckwith-Wiedemann syndrome and epigenetic alter-
GE, Maher ER, Reik W: An association between variants in the IGF2 ations of LIT1 and H19. Am J Hum Genet 2003, 72:156 –160
gene and Beckwith-Wiedemann syndrome: interaction between ge- 48. Bliek J, Gicquel C, Maas S, Gaston V, Le Bouc Y, Mannens M:
notype and epigenotype. Hum Mol Genet 2004, 13:247–255 Epigenotyping as a tool for the prediction of tumor risk and tumor
29. Caspary T, Cleary MA, Perlman EJ, Zhang P, Elledge SJ, Tilghman type in patients with Beckwith-Wiedemann syndrome (BWS). J Pe-
SM: Oppositely imprinted genes p57(Kip2) and igf2 interact in a diatr 2004, 145:796 –799
mouse model for Beckwith-Wiedemann syndrome. Genes Dev 1999, 49. Feil R: Epigenetics: ready for the marks. Nature 2009, 461:359 –360
13:3115–3124 50. Feil R: Epigenetic asymmetry in the zygote and mammalian devel-
30. Feil R, Walter J, Allen ND, Reik W: Developmental control of allelic opment. Int J Dev Biol 2009, 53:191–201
methylation in the imprinted mouse Igf2 and H19 genes. Develop- 51. Li M, Squire JA, Weksberg R: Molecular genetics of Wiedemann-
ment 1994, 120:2933–2943 Beckwith syndrome. Am J Med Genet 1998, 79:253–259
31. Sperandeo MP, Ungaro P, Vernucci M, Pedone PV, Cerrato F, Per- 52. Lam WW, Hatada I, Ohishi S, Mukai T, Joyce JA, Cole TR, Donnai D,
one L, Casola S, Cubellis MV, Bruni CB, Andria G, Sebastio G, Reik W, Schofield PN, Maher ER: Analysis of germline CDKN1C
Riccio A: Relaxation of insulin-like growth factor 2 imprinting and (p57KIP2) mutations in familial and sporadic Beckwith-Wiedemann
discordant methylation at KvDMR1 in two first cousins affected by syndrome (BWS) provides a novel genotype-phenotype correlation.
Beckwith-Wiedemann and Klippel-Trenaunay-Weber syndromes. J Med Genet 1999, 36:518 –523
Am J Hum Genet 2000, 66:841– 847 53. Li M, Squire J, Shuman C, Fei YL, Atkin J, Pauli R, Smith A, Nishikawa
32. Sullivan MJ, Taniguchi T, Jhee A, Kerr N, Reeve AE: Relaxation of J, Chitayat D, Weksberg R: Imprinting status of 11p15 genes in
IGF2 imprinting in Wilms tumours associated with specific changes Beckwith-Wiedemann syndrome patients with CDKN1C mutations.
Genomics 2001, 74:370 –376
in IGF2 methylation. Oncogene 1999, 18:7527–7534
54. Timmerman RJ, DeBaun MR, Feinberg AP, Zehnbauer B: Establish-
33. Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP:
ing thresholds for assessing imprinting abnormalities in Beckwith-
Loss of imprinting in colorectal cancer linked to hypomethylation of
Wiedemann syndrome by methylation hybridization. J Mol Diagn
H19 and IGF2. Cancer Res 2002, 62:6442– 6446
2002, 4:237A
34. Eden S, Constancia M, Hashimshony T, Dean W, Goldstein B, Johnson
55. Riccio A, Sparago A, Verde G, De Crescenzo A, Citro V, Cubellis
AC, Keshet I, Reik W, Cedar H: An upstream repressor element plays
MV, Ferrero GB, Silengo MC, Russo S, Larizza L, Cerrato F: Inherited
a role in Igf2 imprinting. EMBO J 2001, 20:3518 –3525
and Sporadic Epimutations at the IGF2–H19 locus in Beckwith-
35. Constancia M, Dean W, Lopes S, Moore T, Kelsey G, Reik W:
Wiedemann syndrome and Wilms’ tumor. Endocr Dev 2009, 14:1–9
Deletion of a silencer element in Igf2 results in loss of imprinting
56. Maher ER, Afnan M, Barratt CL: Epigenetic risks related to assisted
independent of H19. Nat Genet 2000, 26:203–206
reproductive technologies: epigenetics, imprinting. ART and ice-
36. Murrell A, Heeson S, Bowden L, Constancia M, Dean W, Kelsey G,
bergs? Hum Reprod 2003, 18:2508 –2511
Reik W: An intragenic methylated region in the imprinted Igf2 gene
57. Robertson KD: DNA methylation and human disease. Nat Rev Genet
augments transcription. EMBO Rep 2001, 2:1101–1106
2005, 6:597– 610
37. Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith
58. Li M, Squire JA, Weksberg R: Molecular genetics of Beckwith-
A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, Reik W:
Wiedemann syndrome. Curr Opin Pediatr 1997, 9:623– 629
Placental-specific IGF-II is a major modulator of placental and fetal
59. Schneid H, Vazquez MP, Vacher C, Gourmelen M, Cabrol S, Le
growth. Nature 2002, 417:945–948
Bouc Y: The Beckwith-Wiedemann syndrome phenotype and the
38. Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper risk of cancer. Med Pediatr Oncol 1997, 28:411– 415
PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, 60. Reik W, Brown KW, Slatter RE, Sartori P, Elliott M, Maher ER: Allelic
Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ: A mater- methylation of H19 and IGF2 in the Beckwith-Wiedemann syndrome.
nally methylated CpG island in KvLQT1 is associated with an antisense Hum Mol Genet 1994, 3:1297–1301
paternal transcript and loss of imprinting in Beckwith-Wiedemann syn- 61. Meyer SL: Data analysis for scientists and engineers. New York,
drome. Proc Natl Acad Sci USA 1999, 96:8064 – 8069 Wiley, 1975, p 18
39. Sotelo-Avila C, Gonzalez-Crussi F, Fowler JW: Complete and incom- 62. D’Agostino RB, Stephens MA: Goodness-of-fit techniques. New
plete forms of Beckwith-Wiedemann syndrome: their oncogenic po- York, M. Dekker, 1986, p 390
tential. J Pediatr 1980, 96:47–50 63. Bartlett M: Properties of sufficiency and statistical tests, Proceedings
40. Weng EY, Moeschler JB, Graham JM Jr: Longitudinal observations of the Royal Statistical Society 1937, Series A:268 –282
on 15 children with Wiedemann-Beckwith syndrome. Am J Med 64. Levene H. 1960. Robust tests for equality of variances. In: Olkin, I.,
Genet 1995, 56:366 –373 Ghurye, SG, Hoeffding, W, Madow, WG, and Mann, HB (eds.).
41. Goldman M, Smith A, Shuman C, Caluseriu O, Wei C, Steele L, Ray Contributions to probability and statistics. Essays in honor of Harold
P, Sadowski P, Squire J, Weksberg R, Rosenblum ND: Renal abnor- Hotelling. Stanford University Press, Stanford, Carolina. pp 278 –292
malities in beckwith-wiedemann syndrome are associated with 65. Schumacher A, Doerfler W: Influence of in vitro manipulation on
11p15.5 uniparental disomy. J Am Soc Nephrol 2002, 13:2077–2084 the stability of methylation patterns in the Snurf/Snrpn-imprinting
42. Hoyme HE, Seaver LH, Jones KL, Procopio F, Crooks W, Feingold region in mouse embryonic stem cells. Nucleic Acids Res 2004,
M: Isolated hemihyperplasia (hemihypertrophy): report of a pro- 32:1566 –1576
spective multicenter study of the incidence of neoplasia and review. 66. Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik
Am J Med Genet 1998, 79:274 –278 W, Feil R: Altered imprinted gene methylation and expression in
43. Martin RA, Grange DK, Zehnbauer B, Debaun MR: LIT1 and H19 completely ES cell-derived mouse fetuses: association with aberrant
methylation defects in isolated hemihyperplasia. Am J Med Genet A phenotypes. Development 1998, 125:2273–2282
2005, 134A:129 –131 67. Szabo PE, Mann JR: Biallelic expression of imprinted genes in the
44. Shuman C, Smith AC, Steele L, Ray PN, Clericuzio C, Zackai E, Parisi mouse germ line: implications for erasure, establishment, and mech-
MA, Meadows AT, Kelly T, Tichauer D, Squire JA, Sadowski P, anisms of genomic imprinting. Genes Dev 1995, 9:1857–1868
Weksberg R: Constitutional UPD for chromosome 11p15 in individ- 68. Olek A, Walter J: The pre-implantation ontogeny of the H19 methyl-
uals with isolated hemihyperplasia is associated with high tumor risk ation imprint. Nat Genet 1997, 17:275–276You can also read